Abstract
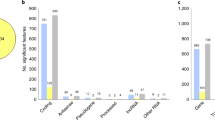
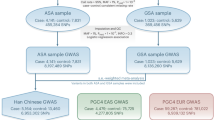
Genetic analyses for bipolar disorder (BD) have achieved prominent success in Europeans in recent years, whereas its genetic basis in other populations remains relatively less understood. We herein report that the leading risk locus for BD in European genome-wide association studies (GWAS), the single-nucleotide polymorphism (SNP) rs9834970 near TRANK1 at 3p22 region, is also genome-wide significantly associated with BD in a meta-analysis of four independent East Asian samples including 5748 cases and 65,361 controls (p = 2.27 × 10−8, odds ratio = 1.136). Expression quantitative trait loci (eQTL) analyses and summary data-based Mendelian randomization (SMR) analyses in multiple human brain samples suggest that lower TRANK1 mRNA expression is a principal BD risk factor explaining its genetic risk signals at 3p22. We also identified another SNP rs4789 in the 3′ untranslated region (3′UTR) of TRANK1 showing stronger eQTL associations as well as genome-wide significant association with BD. Despite the relatively unclear neuronal function of TRANK1, our mRNA expression analyses in the human brains and in rat primary cortical neurons reveal that genes highly correlated with TRANK1 are significantly enriched in the biological processes related to dendritic spine, synaptic plasticity, axon guidance and circadian entrainment, and are also more likely to exhibit strong associations in psychiatric GWAS (e.g., the CACNA1C gene). Overall, our results support that TRANK1 is a potential BD risk gene. Further studies elucidating its roles in this illness are needed.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Vieta E, Berk M, Schulze TG, Carvalho AF, Suppes T, Calabrese JR, et al. Bipolar disorders. Nat Rev Dis Prim. 2018;4:18008.
Gordovez FJA, McMahon FJ. The genetics of bipolar disorder. Mol Psychiatry. 2020;25:544–59.
Zhang C, Xiao X, Li T, Li M. Translational genomics and beyond in bipolar disorder. Mol Psychiatry. 2020. https://doi.org/10.1038/s41380-41020-40782-41389.
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803.
Cichon S, Muhleisen TW, Degenhardt FA, Mattheisen M, Miro X, Strohmaier J, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88:372–81.
Hou L, Bergen SE, Akula N, Song J, Hultman CM, Landen M, et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet. 2016;25:3383–94.
Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJ, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry. 2013;18:195–205.
Muhleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun. 2014;5:3339.
Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–8.
Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–83.
Ikeda M, Takahashi A, Kamatani Y, Okahisa Y, Kunugi H, Mori N, et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol Psychiatry. 2018;23:639–47.
Zhao L, Chang H, Zhou DS, Cai J, Fan W, Tang W, et al. Replicated associations of FADS1, MAD1L1, and a rare variant at 10q26.13 with bipolar disorder in Chinese population. Transl Psychiatry. 2018;8:270.
Wang L, Liu W, Li X, Xiao X, Li L, Liu F, et al. Further evidence of an association between NCAN rs1064395 and bipolar disorder. Mol Neuropsychiatry. 2018;4:30–4.
Goes FS, Hamshere ML, Seifuddin F, Pirooznia M, Belmonte-Mahon P, Breuer R, et al. Genome-wide association of mood-incongruent psychotic bipolar disorder. Transl Psychiatry. 2012;2:e180.
Lee MT, Chen CH, Lee CS, Chen CC, Chong MY, Ouyang WC, et al. Genome-wide association study of bipolar I disorder in the Han Chinese population. Mol Psychiatry. 2011;16:548–56.
Jiang X, Detera-Wadleigh SD, Akula N, Mallon BS, Hou L, Xiao T, et al. Sodium valproate rescues expression of TRANK1 in iPSC-derived neural cells that carry a genetic variant associated with serious mental illness. Mol Psychiatry. 2019;24:613–24.
Li W, Yang Y, Luo B, Zhang Y, Song X, Li M, et al. Association of SYNE1 locus with bipolar disorder in Chinese population. Hereditas. 2019;156:19.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9.
Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53.
Ng B, White CC, Klein HU, Sieberts SK, McCabe C, Patrick E, et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat Neurosci. 2017;20:1418–26.
GTEx Consortium, Laboratory Data Analysis, Coordinating Center Analysis Working Group, Statistical Methods groups-Analysis Working Group, Enhancing GTEx groups, NIH Common Fund, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13.
Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;6:e1000770.
Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–7.
Wu Y, Zeng J, Zhang F, Zhu Z, Qi T, Zheng Z, et al. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat Commun. 2018;9:918.
Li HJ, Qu N, Hui L, Cai X, Zhang CY, Zhong BL, et al. Further confirmation of netrin 1 receptor (DCC) as a depression risk gene via integrations of multi-omics data. Transl Psychiatry. 2020;10:98.
Wang Z, Wang N, Li Z, Xiao F, Dai J. Human high intelligence is involved in spectral redshift of biophotonic activities in the brain. Proc Natl Acad Sci USA. 2016;113:8753–8.
Li H, Chang H, Song X, Liu W, Li L, Wang L, et al. Integrative analyses of major histocompatibility complex loci in the genome-wide association studies of major depressive disorder. Neuropsychopharmacology. 2019;44:1552–61.
Liu W, Yan H, Zhou D, Cai X, Zhang Y, Li S, et al. The depression GWAS risk allele predicts smaller cerebellar gray matter volume and reduced SIRT1 mRNA expression in Chinese population. Transl Psychiatry. 2019;9:333.
Cai X, Yang ZH, Li HJ, Xiao X, Li M, Chang H. A human-specific schizophrenia risk tandem repeat affects alternative splicing of a human-unique isoform AS3MTd2d3 and mushroom dendritic spine density. Schizophr Bull. 2020. https://doi.org/10.1093/schbul/sbaa1098.
PsychEncode Consortium, Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, et al. The PsychENCODE project. Nat Neurosci. 2015;18:1707–12.
Li H, Zhou DS, Chang H, Wang L, Liu W, Dai SX, et al. Interactome analyses implicated CAMK2A in the genetic predisposition and pharmacological mechanism of bipolar disorder. J Psychiatr Res. 2019;115:165–75.
Yang Z, Zhou D, Li H, Cai X, Liu W, Wang L, et al. The genome-wide risk alleles for psychiatric disorders at 3p21.1 show convergent effects on mRNA expression, cognitive function and mushroom dendritic spine. Mol Psychiatry. 2020;25:48–66.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–8.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–97.
Gallagher MD, Chen-Plotkin AS. The post-GWAS era: from association to function. Am J Hum Genet. 2018;102:717–30.
Gonzalez SD, Xu C, Ramirez ME, Zavala JM, Armas R, Contreras SA, et al. Family-based association of an ANK3 haplotype with bipolar disorder in Latino populations. Transl Psychiatry. 2013;3:e265.
Liu J, Li ZQ, Li JY, Li T, Wang T, Li Y, et al. Polymorphisms and haplotypes in the YWHAE gene increase susceptibility to bipolar disorder in Chinese Han population. J Clin Psychiatry. 2012;73:e1276–82.
Nurnberger JI Jr., Koller DL, Jung J, Edenberg HJ, Foroud T, Guella I, et al. Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry. 2014;71:657–64.
Akula N, Wendland JR, Choi KH, McMahon FJ. An integrative genomic study implicates the postsynaptic density in the pathogenesis of bipolar disorder. Neuropsychopharmacology. 2016;41:886–95.
Konopaske GT, Lange N, Coyle JT, Benes FM. Prefrontal cortical dendritic spine pathology in schizophrenia and bipolar disorder. JAMA Psychiatry. 2014;71:1323–31.
Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–93.
Forrest MP, Parnell E, Penzes P. Dendritic structural plasticity and neuropsychiatric disease. Nat Rev Neurosci. 2018;19:215–34.
Focking M, Dicker P, Lopez LM, Hryniewiecka M, Wynne K, English JA, et al. Proteomic analysis of the postsynaptic density implicates synaptic function and energy pathways in bipolar disorder. Transl Psychiatry. 2016;6:e959.
Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–9.
Pardinas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–9.
Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81.
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219.
Jaffe AE, Straub RE, Shin JH, Tao R, Gao Y, Collado-Torres L, et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci. 2018;21:1117–25.
Acknowledgements
We acknowledge with appreciation all the individuals with bipolar disorders and healthy controls whose contributions made this work possible. We are deeply grateful to all the participants working on this project. The authors wish to acknowledge Dr. Bernard Ng (Departments of Statistics and Medical Genetics, University of British Columbia, Vancouver, BC, Canada), Dr. Sara Mostafavi (Departments of Statistics and Medical Genetics, University of British Columbia, Vancouver, BC, Canada), and Dr. Philip L De Jager (Broad Institute, Cambridge, MA, USA) for providing the individual-level TRANK1 expression and genotypes in the Brain xQTL dataset.
Author information
Authors and Affiliations
Contributions
H.C., L.L., and M.L. conceived the study and interpreted the results. W.L., X.C., M.S., H.-J.L., L.Z., W.L., and L.W. performed the DNA extraction, SNP genotyping, statistical analysis, and the primary experiments. H.-J.L. and C.-Y.Z. performed the RNA-seq and pathway analysis. W.L., M.S., Y.Y., L.Z., M.S., Y.Z., C.Z., D.-S.Z., X.L., L.H., Q.-F.J., N.Q., B.-L.Z., S.-F.Z., J.C., B.X., Y.L., X.S., W.F., W.T., W.T., J.T., X.C., Y.F., and L.L. carried out subject recruitment and phenotype analysis. W.Y. and D.Z. contributed to design of sample analysis. X.X., H.C., L.L., and M.L. drafted the manuscript, and all authors contributed to the final version of the paper.
Corresponding authors
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, W., Cai, X., Li, HJ. et al. Independent replications and integrative analyses confirm TRANK1 as a susceptibility gene for bipolar disorder. Neuropsychopharmacol. 46, 1103–1112 (2021). https://doi.org/10.1038/s41386-020-00788-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41386-020-00788-4
This article is cited by
-
Permethrin exposure primes neuroinflammatory stress response to drive depression-like behavior through microglial activation in a mouse model of Gulf War Illness
Journal of Neuroinflammation (2024)
-
TrkB-dependent regulation of molecular signaling across septal cell types
Translational Psychiatry (2024)
-
Sleep Disorder Kleine–Levin Syndrome (KLS) Joins the List of Polygenic Brain Disorders Associated with Obstetric Complications
Cellular and Molecular Neurobiology (2023)
-
Multi-ancestry meta-analysis and fine-mapping in Alzheimer’s disease
Molecular Psychiatry (2023)
-
Cell-type-specific cis-eQTLs in eight human brain cell types identify novel risk genes for psychiatric and neurological disorders
Nature Neuroscience (2022)