Fig. 1
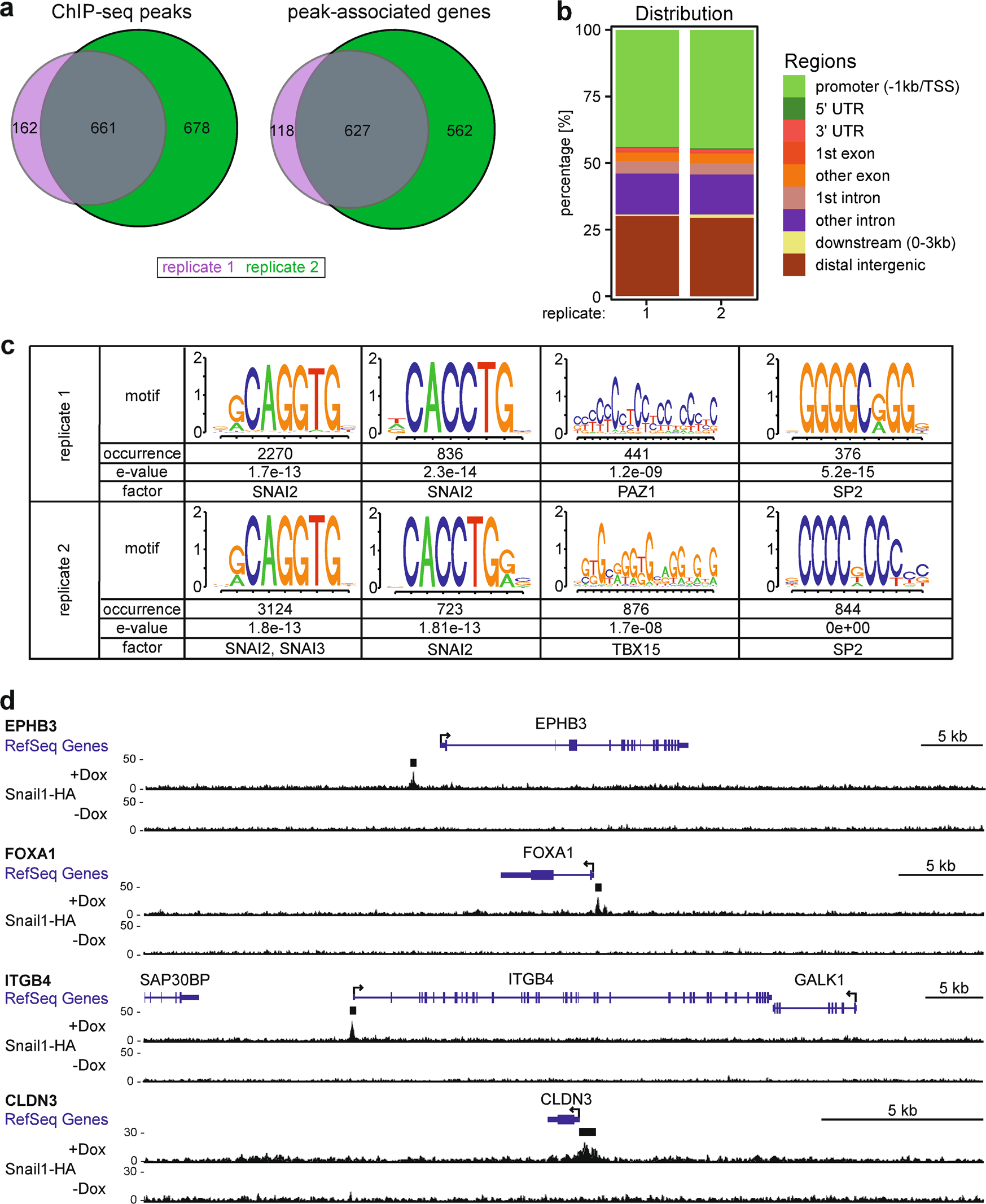
Genome-wide identification of Snail1-HA DNA-binding sites in LS174T CRC cells. a Venn diagrams showing numbers of distinct and common ChIP-seq peaks and peak-associated genes from two independent experiments with LS174T cells stably transduced with retroviral expression vectors for Dox-inducible Snail1-HA. Cells were treated with 0.1 μg ml−1 Dox for 6 h prior to harvest and processing. b Distribution of Snail1-HA ChIP-seq peaks in replicates 1 and 2 across genomic regions. c Most abundant DNA sequence motifs identified in Snail1-HA ChIP-seq peaks from both replicates. Frequencies of motif occurrence, alignment quality (e-value), and transcription factors potentially recognizing the motifs are displayed. d Genome browser views of the indicated RefSeq gene loci depicting ChIP-seq tracks with called peaks (black bars) for Snail1-HA in LS174T cells in the presence or absence of Dox. Tracks represent a combination of replicates 1 and 2 and are based on hg19 sequence information
