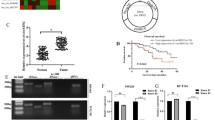
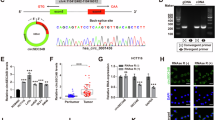
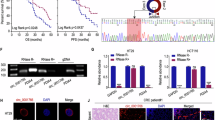
Abstract
Tumor progression of colorectal cancer (CRC) seriously affects patient prognosis. For CRC patients with advanced-stage disease, it is still necessary to continuously explore more effective targeted therapeutic drugs. Circular RNAs (circRNAs) are involved in the regulation of tumor biology. We screened circAURKA, which was significantly highly expressed in CRC by previous high-throughput RNA sequencing. In vitro experiments were performed to investigate the effect of the circRNA on the proliferation and metastasis of HCT116 and SW480 cells. In addition, we used the EdU assay, Transwell assay, nude mouse xenograft tumor model and nude mouse tail vein metastasis model to examine the effect of circAURKA on the proliferation and metastasis of CRC. Mechanistically, fluorescent in situ hybridization (FISH), RNA pull-down, RNA immunoprecipitation (RIP), protein coimmunoprecipitation (co-IP) experiments and animal models were performed to confirm the underlying mechanisms of circAURKA. CircAURKA was significantly highly expressed in CRC tissues and colorectal cells and mainly present in the cytoplasm. The circRNA promoted the proliferation and metastasis of CRC cells in vitro and in vivo. In terms of the molecular mechanism, circAURKA inhibited the degradation of the CTNNB1 protein by promoting the interaction between ACLY and the CTNNB1 protein, thereby promoting the proliferation and metastasis of CRC cells. In addition, circAURKA stability was regulated by m6A methylation modification. This study revealed that circAURKA promoted the proliferation and metastasis of CRC by inhibiting CTNNB1 protein degradation, providing a basis for the development of targeted drugs to control CRC progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All the data used in the current study are available from the corresponding authors upon reasonable request.
References
Collaborators GBDCC. The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2019;4:913–33.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA: Cancer J Clin. 2021;71:209–49.
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–80.
Eng C, Hochster H. Early-onset colorectal cancer: the mystery remains. J Natl Cancer Inst. 2021;113:1608–10.
Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–8.
Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–74.
Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA. 2021;325:669–85.
Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206.
Long F, Lin Z, Li L, Ma M, Lu Z, Jing L, et al. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol Cancer. 2021;20:26.
Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6:319–36.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526.
Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503–17.
Liu CX, Chen LL. Circular RNAs: characterization, cellular roles, and applications. Cell. 2022;185:2016–34.
Peng Y, Xu Y, Zhang X, Deng S, Yuan Y, Luo X, et al. A novel protein AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin signaling pathway to promote gastric cancer progression. Mol Cancer. 2021;20:158.
Xue C, Li G, Zheng Q, Gu X, Bao Z, Lu J, et al. The functional roles of the circRNA/Wnt axis in cancer. Mol Cancer. 2022;21:108.
Zhao H, Ming T, Tang S, Ren S, Yang H, Liu M, et al. Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol Cancer. 2022;21:144.
Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–20.
Liu C, Wang L, Liu X, Tan Y, Tao L, Xiao Y, et al. Cytoplasmic SHMT2 drives the progression and metastasis of colorectal cancer by inhibiting β-catenin degradation. Theranostics. 2021;11:2966–86.
Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–804.
Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, et al. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–10.
Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–47.
Cselenyi CS, Jernigan KK, Tahinci E, Thorne CA, Lee LA, Lee E. LRP6 transduces a canonical Wnt signal independently of Axin degradation by inhibiting GSK3’s phosphorylation of beta-catenin. Proc Natl Acad Sci USA. 2008;105:8032–7.
Kim SE, Huang H, Zhao M, Zhang X, Zhang A, Semonov MV, et al. Wnt stabilization of beta-catenin reveals principles for morphogen receptor-scaffold assemblies. Science. 2013;340:867–70.
Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, et al. Wnt signaling through inhibition of beta-catenin degradation in an intact Axin1 complex. Cell. 2012;149:1245–56.
Schuijers J, Mokry M, Hatzis P, Cuppen E, Clevers H. Wnt-induced transcriptional activation is exclusively mediated by TCF/LEF. EMBO J. 2014;33:146–56.
Guo Y, Guo Y, Chen C, Fan D, Wu X, Zhao L, et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer. 2021;20:93.
Patel SG, Karlitz JJ, Yen T, Lieu CH, Boland CR. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol Hepatol. 2022;7:262–74.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91.
Huang D, Zhu X, Ye S, Zhang J, Liao J, Zhang N, et al. Tumour circular RNAs elicit anti-tumour immunity by encoding cryptic peptides. Nature. 2024;625:593–602.
Wei G, Chen X, Ruan T, Ma X, Zhu X, Wen W, et al. Human gastric cancer progression and stabilization of ATG2B through RNF5 binding facilitated by autophagy-associated CircDHX8. Cell Death Dis. 2024;15:410.
Chen B, Scurrah CR, McKinley ET, Simmons AJ, Ramirez-Solano MA, Zhu X, et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell. 2021;184:6262–6280.e6226.
Gil-Raga M, Jantus-Lewintre E, Gallach S, Giner-Bosch V, Frangi-Caregnato A, Safont-Aguilera MJ, et al. Molecular subtypes in early colorectal cancer associated with clinical features and patient prognosis. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of. Natl Cancer Inst Mexico. 2018;20:1422–9.
Liao Y, Feng J, Sun W, Wu C, Li J, Jing T, et al. CIRP promotes the progression of non-small cell lung cancer through activation of Wnt/β-catenin signaling via CTNNB1. J Exp Clin Cancer Res. 2021;40:275.
Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480:118–22.
Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang X, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7:3.
Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–42.
Wang L, Li B, Yi X, Xiao X, Zheng Q, Ma L. Circ_SMAD4 promotes gastric carcinogenesis by activating wnt/β-catenin pathway. Cell Prolif. 2021;54:e12981.
Zhou Q, Fu Q, Shaya M, Kugeluke Y, Li S, Dilimulati Y. Knockdown of circ_0055412 promotes cisplatin sensitivity of glioma cells through modulation of CAPG and Wnt/β-catenin signaling pathway. CNS Neurosci Ther. 2022;28:884–96.
Funding
This study was supported by The National Natural Science Foundation of China (82173055), Scientific Research and Innovation Team of The First Affiliated Hospital of Zhengzhou University (ZYCXTD2023017), The Provincial and Ministry Co-constructed Key Projects of Henan Medical Science and Technology (SBGJ202102134), Henan Provincial Health and Health Commission Joint Construction Project (LHGJ20200158), and Henan Province Young and Middle-aged Health Science and Technology Innovation Leading Talent Project (YXKC2022016).
Author information
Authors and Affiliations
Contributions
ZS, YL, and JH provided direction and guidance throughout the preparation of this manuscript. PD, YG, SL, YX, HS, WW, and CC performed all experiments in this study. PD, YG, SL, and SH performed the data analysis and wrote the manuscript. JL and ZJ collected the related references and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The authors declare that all methods were performed in accordance with all relevant guidelines and regulations. The human cancer tissues used in this study were approved by Ethnics Committee of The First Affiliated Hospital of Zhengzhou University (2019-KY-423). The Zhengzhou University Institutional Animal Care and Use Committee reviewed and approved all animal research protocols.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, Z., Dang, P., Guo, Y. et al. Targeting CircAURKA prevents colorectal cancer progression via enhancing CTNNB1 protein degradation. Oncogene 43, 3388–3401 (2024). https://doi.org/10.1038/s41388-024-03155-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-024-03155-5