Abstract
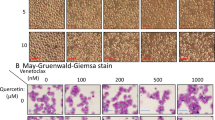
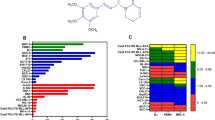
Acute myeloid leukemia (AML) represents the most prevalent malignancy within the hematologic system, characterized by refractory relapses and a scarcity of effective treatment options. Karyopherin subunit beta-1 (KPNB1) is a member of karyopherin β family, mediating the nuclear import of its cargoes. In this study, we found that elevated expression levels of KPNB1 are associated with unfavorable outcomes in patients with AML. The knockdown of KPNB1 resulted in growth inhibition and apoptosis in AML cells. Additionally, pharmacological inhibition of KPNB1 using the specific inhibitor importazole (IPZ) significantly reduced tumor burden and prolonged survival in MLL-AF9-induced AML mice. Notably, the inhibition of KPNB1 by IPZ significantly enhanced the sensitivity of both AML cell lines and patient-derived cells to venetoclax in vitro and in xenograft mice models. At the molecular level, we identified an unrecognized cargo of KPNB1, high mobility group 2 (HMGB2), which plays a crucial role in DNA damage repair. Inhibition of KPNB1 resulted in impaired nuclear import of HMGB2, eventually leading to compromised DNA damage repair in AML cells. Overall, our findings elucidate the essential roles of KPNB1 in AML cells through the HMGB2-DNA damage repair axis and highlight a promising therapeutic target for AML intervention.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Supplemental Figures and Tables are available in the Supplemental Figures and Tables file. RNA-seq data were deposited in the National Center for Biotechnology Information GEO database and are available under accession number GSE277912. Proteomics are available at PRIDE-Proteomics Identification Database under accession numbers PXD056172. All other data are available in the article and its supplementary materials and are also available upon request from the corresponding author.
References
Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–52.
Carter JL, Hege K, Yang J, Kalpage HA, Su Y, Edwards H, et al. Targeting multiple signaling pathways: the new approach to acute myeloid leukemia therapy. Signal Transduct Target Ther. 2020;5:288.
Dombret H, Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2016;127:53–61.
Dohner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–77.
Ding B, Sepehrimanesh M. Nucleocytoplasmic transport: regulatory mechanisms and the implications in neurodegeneration. Int J Mol Sci. 2021;22:4165.
Wing CE, Fung HYJ, Chook YM. Karyopherin-mediated nucleocytoplasmic transport. Nat Rev Mol Cell Biol. 2022;23:307–28.
Azmi AS, Khan HY, Muqbil I, Aboukameel A, Neggers JE, Daelemans D, et al. Preclinical assessment with clinical validation of selinexor with gemcitabine and nab-paclitaxel for the treatment of pancreatic ductal adenocarcinoma. Clin Cancer Res. 2020;26:1338–48.
Syed YY. Selinexor: first global approval. Drugs. 2019;79:1485–94.
Pan L, Cheng C, Duan P, Chen K, Wu Y, Wu Z. XPO1/CRM1 is a promising prognostic indicator for neuroblastoma and represented a therapeutic target by selective inhibitor verdinexor. J Exp Clin Cancer Res. 2021;40:255.
Verbeke D, Demeyer S, Prieto C, de Bock CE, De Bie J, Gielen O, et al. The XPO1 inhibitor KPT-8602 synergizes with dexamethasone in acute lymphoblastic leukemia. Clin Cancer Res. 2020;26:5747–58.
Chari A, Vogl DT, Gavriatopoulou M, Nooka AK, Yee AJ, Huff CA, et al. Oral selinexor-dexamethasone for triple-class refractory multiple myeloma. N Engl J Med. 2019;381:727–38.
Deng M, Zhang M, Xu-Monette ZY, Pham LV, Tzankov A, Visco C, et al. XPO1 expression worsens the prognosis of unfavorable DLBCL that can be effectively targeted by selinexor in the absence of mutant p53. J Hematol Oncol. 2020;13:148.
Shi Q, Lin M, Cheng X, Zhang Z, Deng S, Lang K, et al. KPNB1-mediated nuclear import in cancer. Eur J Pharm. 2023;955:175925.
Cingolani G, Petosa C, Weis K, Müller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–9.
Lee SJ, Matsuura Y, Liu SM, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–6.
Chook YM, Süel KE. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–606.
Matsuura Y. Mechanistic insights from structural analyses of Ran-GTPase-driven nuclear export of proteins and RNAs. J Mol Biol. 2016;428:2025–39.
Ha S, Jeong J, Oh J, Rhee S, Ham SW. Inhibition of the nuclear transporter, Kpnβ1, results in prolonged mitotic arrest and activation of the intrinsic apopesulting in repression of cancer invasion. ChemBioChem. 2018;19:131–5.
Du W, Zhu J, Zeng Y, Liu T, Zhang Y, Cai T, et al. KPNB1-mediated nuclear translocation of PD-L1 promotes non-small cell lung cancer cell proliferation via the Gas6/MerTK signaling pathway. Cell Death Differ. 2021;28:1284–300.
van der Watt PJ, Maske CP, Hendricks DT, Parker MI, Denny L, Govender D, et al. The Karyopherin proteins, Crm1 and Karyopherin beta1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Int J Cancer. 2009;124:1829–40.
Yang J, Guo Y, Lu C, Zhang R, Wang Y, Luo L, et al. Inhibition of Karyopherin beta 1 suppresses prostate cancer growth. Oncogene. 2019;38:4700–14.
Wang T, Huang Z, Huang N, Peng Y, Gao M, Wang X, et al. Inhibition of KPNB1 inhibits proliferation and promotes apoptosis of chronic myeloid leukemia cells through regulation of E2F1. Onco Targets Ther. 2019;12:10455–67.
Kodama M, Kodama T, Newberg JY, Katayama H, Kobayashi M, Hanash SM, et al. In vivo loss-of-function screens identify KPNB1 as a new druggable oncogene in epithelial ovarian cancer. Proc Natl Acad Sci USA. 2017;114:E7301–10.
Kim YH, Ha S, Kim J, Ham SW. Identification of KPNB1 as a cellular target of aminothiazole derivatives with anticancer activity. ChemMedChem. 2016;11:1406–9.
Yoshino H, Sato Y, Nakano M. KPNB1 inhibitor importazole reduces ionizing radiation-increased cell surface PD-L1 expression by modulating expression and nuclear import of IRF1. Curr Issues Mol Biol. 2021;43:153–62.
Kelenis DP, Rodarte KE, Kollipara RK, Pozo K, Choudhuri SP, Spainhower KB, et al. Inhibition of karyopherin β1-mediated nuclear import disrupts oncogenic lineage-defining transcription factor activity in small cell lung cancer. Cancer Res. 2022;82:3058–73.
Hintersteiner M, Ambrus G, Bednenko J, Schmied M, Knox AJ, Meisner NC, et al. Identification of a small molecule inhibitor of importin β mediated nuclear import by confocal on-bead screening of tagged one-bead one-compound libraries. ACS Chem Biol. 2010;5:967–79.
Chen Z, Guo Q, Huang S, Li L, Wu F, Liu Z, et al. Overcoming adaptive resistance in AML by synergistically targeting FOXO3A-GNG7-mTOR axis with FOXO3A inhibitor Gardenoside and rapamycin. Genes Dis. 2023;11:397–412.
Chen Z, Huo D, Li L, Liu Z, Li Z, Xu S, et al. Nuclear DEK preserves hematopoietic stem cells potential via NCoR1/HDAC3-Akt1/2-mTOR axis. J Exp Med. 2021;218:e20201974.
Lanevski A, Giri AK, Aittokallio T. SynergyFinder 3.0: an interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic Acids Res. 2022;50:W739–43.
Uddin MH, Zonder JA, Azmi AS. Exportin 1 inhibition as antiviral therapy. Drug Discov Today. 2020;25:1775–81.
Angus L, van der Watt PJ, Leaner VD. Inhibition of the nuclear transporter, Kpnβ1, results in prolonged mitotic arrest and activation of the intrinsic apoptotic pathway in cervical cancer cells. Carcinogenesis. 2014;35:1121–31.
Waga S, Mizuno S, Yoshida M. Nonhistone protein HMG1 removes the transcriptional block caused by left-handed Z-form segment in a supercoiled DNA. Biochem Biophys Res Commun. 1988;153:334–9.
Bianchi ME, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–9.
Pearson CE, Ruiz MT, Price GB, Zannis-Hadjopoulos M. Cruciform DNA binding protein in HeLa cell extracts. Biochemistry. 1994;33:14185–96.
Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, et al. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol Cell. 2018;70:730–44.
Monte E, Rosa-Garrido M, Karbassi E, Chen H, Lopez R, Rau CD, et al. Reciprocal regulation of the cardiac epigenome by chromatin structural proteins Hmgb and Ctcf: implications for transcriptional regulation. J Biol Chem. 2016;291:15428–46.
Brambilla F, Garcia-Manteiga JM, Monteleone E, Hoelzen L, Zocchi A, Agresti A, et al. Nucleosomes effectively shield DNA from radiation damage in living cells. Nucleic Acids Res. 2020;48:8993–9006.
Han Y, Wang X. The emerging roles of KPNA2 in cancer. Life Sci. 2020;241:117140.
Lee Y, Jang AR, Francey LJ, Sehgal A, Hogenesch JB. KPNB1 mediates PER/CRY nuclear translocation and circadian clock function. Elife. 2015;4:e08647.
Lam MH, Briggs LJ, Hu W, Martin TJ, Gillespie MT, Jans DA. Importin beta recognizes parathyroid hormone-related protein with high affinity and mediates its nuclear import in the absence of importin alpha. J Biol Chem. 1999;274:7391–8.
Starkova T, Polyanichko A, Tomilin AN, Chikhirzhina E. Structure and functions of HMGB2 protein. Int J Mol Sci. 2023;24:8334.
Wheeler EC, Martin B, Doyle WC, Neaher S, Conway CA, Pitton CN, et al. Splicing modulators impair DNA damage response and induce killing of cohesin-mutant MDS and AML. Sci Transl Med. 2024;16:eade2774.
Huang R, Zhou PK. DNA damage repair: historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct Target Ther. 2021;6:254.
Watanabe F, Shirakawa H, Yoshida M, Tsukada K, Teraoka H. Stimulation of DNA-dependent protein kinase activity by high mobility group proteins 1 and 2. Biochem Biophys Res Commun. 1994;202:736–42.
Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell. 2017;66:801–17.
Redon CE, Nakamura AJ, Martin OA, Parekh PR, Weyemi US, Bonner WM. Recent developments in the use of γ-H2AX as a quantitative DNA double-strand break biomarker. Aging. 2011;3:168–74.
Carbonell B, Álvarez J, Santa-González GA, Delgado JP. COMET assay for detection of DNA damage during axolotl tail regeneration. Methods Mol Biol. 2023;2562:183–94.
Killock D. Venetoclax in AML: efficacy confirmed. Nat Rev Clin Oncol. 2020;17:592.
Nachmias B, Aumann S, Haran A, Schimmer AD. Venetoclax resistance in acute myeloid leukaemia—clinical and biological insights. Br J Haematol. 2024;204:1146–58.
van der Watt PJ, Chi A, Stelma T, Stowell C, Strydom E, Carden S, et al. Targeting the nuclear import receptor Kpnβ1 as an anticancer therapeutic. Mol Cancer Ther. 2016;15:560–73.
Chi RA, van der Watt P, Wei W, Birrer MJ, Leaner VD. Inhibition of Kpnβ1 mediated nuclear import enhances cisplatin chemosensitivity in cervical cancer. BMC Cancer. 2021;21:106.
Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–13.
Fan Z, Beresford PJ, Zhang D, Lieberman J. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol Cell Biol. 2002;22:2810–20.
Ouellet V, Le Page C, Guyot MC, Lussier C, Tonin PN, Provencher DM, et al. SET complex in serous epithelial ovarian cancer. Int J Cancer. 2006;119:2119–26.
Mathai C, Jourd’Heuil F, Pham L, Gilliard K, Howard D, Balnis J, et al. Regulation of DNA damage and transcriptional output in the vasculature through a cytoglobin-HMGB2 axis. Redox Biol. 2023;65:102838.
Newell S, van der Watt PJ, Leaner VD. Therapeutic targeting of nuclear export and import receptors in cancer and their potential in combination chemotherapy. IUBMB Life. 2024;76:4–25.
Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M, et al. Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem Biol. 2011;6:700–8.
Funding
This work was supported by grants from Chongqing Science Fund for Distinguished Young Scholars (CSTB2022NSCQ-JQX0032), CQMU Program for Youth Innovation in Future Medicine (W0156), Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No. KJQN202400465), and Chongqing Postdoctoral Science Foundation (CSTB2023NSCQ-BHX0144 and 2022CQBSHTB1012).
Author information
Authors and Affiliations
Contributions
Y Hou, Z Chen, and H Zeng conceived the project, analyzed data, and revised the paper. Y Xie performed experiments, analyzed data, and wrote the paper. R Zhao, Y Zheng, Y Li, F Wu, Y Lei, and L Li contributed to the data analysis and paper revision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The authors affirm that all methods were conducted in accordance with the relevant guidelines and regulations. All mice were housed at the Animal Center of Chongqing Medical University, and approval was obtained from the Animal Committee of the Institute of Zoology at Chongqing Medical University (reference number: IACUC-CQMU-2024-0046, “The study on pathogenesis and targeted intervention of acute myeloid leukemia”). Whole BM samples were obtained from AML patients after informed consent of sample use for research. The use of human samples was approved by the Medical Ethics Committees of Chongqing Medical University (reference number: 2024019, “The study on pathogenesis and targeted intervention of acute myeloid leukemia”).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xie, Y., Zhao, R., Zheng, Y. et al. Targeting KPNB1 suppresses AML cells by inhibiting HMGB2 nuclear import. Oncogene 44, 1646–1661 (2025). https://doi.org/10.1038/s41388-025-03340-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03340-0