Abstract
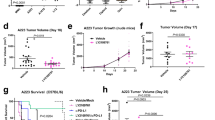
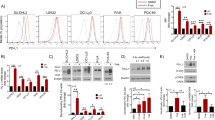
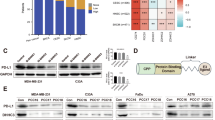
Blockade of immune checkpoints, such as programmed death-ligand 1 (PD-L1), has shown promise in cancer treatment; however, clinical response remains limited in many cancer types. Our previous research demonstrated that p300/CBP mediates the acetylation of the PD-L1 promoter, regulating PD-L1 expression. In this study, we further investigated the role of the p300/CBP bromodomain in regulating PD-L1 expression using CCS1477, a selective bromodomain inhibitor developed by our team. We found that the p300/CBP bromodomain is essential for H3K27 acetylation at PD-L1 enhancers. Inhibiting this modification significantly reduced enhancer activity and PD-L1 transcription, including exosomal PD-L1, which has been implicated as key contributors to resistance against PD-L1 blockade therapy in various cancers. Furthermore, CCS1477 treatment resulted in a marked reduction of myeloid-derived suppressor cells (MDSCs) in the tumor microenvironment (TME) by inhibiting key cytokines such as IL6, CSF1, and CSF2, which are crucial for MDSC differentiation and recruitment. By reducing PD-L1 expression and modulating the immunosuppressive TME, CCS1477 creates a more favorable environment for tumor-infiltrating lymphocytes, significantly enhancing the efficacy of immune checkpoint blockade (ICB) therapy. Notably, these effects were observed in both prostate cancer and melanoma models, underscoring the broad therapeutic potential of p300/CBP bromodomain inhibition in improving ICB outcomes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting the findings of this study are either included in the manuscript or available from the corresponding authors upon reasonable request.
References
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48.
Swami U, McFarland TR, Nussenzveig R, Agarwal N. Advanced prostate cancer: treatment advances and future directions. Trends Cancer. 2020;6:702–15.
Chen Y, Zhou Q, Hankey W, Fang X, Yuan F. Second generation androgen receptor antagonists and challenges in prostate cancer treatment. Cell Death Dis. 2022;13:632.
Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125:3384–91.
Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61.
Sharma P, Pachynski RK, Narayan V, Flechon A, Gravis G, Galsky MD, et al. Nivolumab plus ipilimumab for metastatic castration-resistant prostate cancer: preliminary analysis of patients in the CheckMate 650 trial. Cancer Cell. 2020;38:489–99.e483.
Liu J, He D, Cheng L, Huang C, Zhang Y, Rao X, et al. p300/CBP inhibition enhances the efficacy of programmed death-ligand 1 blockade treatment in prostate cancer. Oncogene. 2020;39:3939–51.
Lu X, Horner JW, Paul E, Shang X, Troncoso P, Deng P, et al. Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature. 2017;543:728–32.
Wang J, Zhang HL, Sun X, et al. Exosomal PD-L1 and N-cadherin predict pulmonary metastasis progression for osteosarcoma patients. J Nanobiotechnol. 2020;18:151. https://doi.org/10.1186/s12951-020-00710-6.
Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–6.
Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, et al. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell. 2019;177:414–27.e413.
Wang G, Lu X, Dey P, Deng P, Wu CC, Jiang S, et al. Targeting YAP-dependent MDSC infiltration impairs tumor progression. Cancer Discov. 2016;6:80–95.
Zhao D, Cai L, Lu X, Liang X, Li J, Chen P, et al. Chromatin regulator CHD1 remodels the immunosuppressive tumor microenvironment in PTEN-deficient. Prostate Cancer Cancer Discov. 2020;10:1374–87.
Hossain DM, Pal SK, Moreira D, Duttagupta P, Zhang Q, Won H, et al. TLR9-Targeted STAT3 silencing abrogates immunosuppressive activity of myeloid-derived suppressor cells from prostate cancer patients. Clin Cancer Res. 2015;21:3771–82.
Jin L, Garcia J, Chan E, de la Cruz C, Segal E, Merchant M, et al. Therapeutic targeting of the CBP/p300 bromodomain blocks the growth of castration-resistant prostate cancer. Cancer Res. 2017;77:5564–75.
Zhong J, Ding L, Bohrer LR, Pan Y, Liu P, Zhang J, et al. p300 acetyltransferase regulates androgen receptor degradation and PTEN-deficient prostate tumorigenesis. Cancer Res. 2014;74:1870–80.
Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 2017;550:128–32.
Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, et al. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275:20853–60.
Welti J, Sharp A, Brooks N, Yuan W, McNair C, Chand SN, et al. Targeting the p300/CBP axis in lethal. Prostate Cancer Cancer Discov. 2021;11:1118–37.
Nicosia L, Spencer GJ, Brooks N, Amaral FMR, Basma NJ, Chadwick JA, et al. Therapeutic targeting of EP300/CBP by bromodomain inhibition in hematologic malignancies. Cancer Cell. 2023;41:2136–53.e2113.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Liu J, Zhao Y, He D, Jones KM, Tang S, Allison DB, et al. A kinome-wide CRISPR screen identifies CK1alpha as a target to overcome enzalutamide resistance of prostate cancer. Cell Rep Med. 2023;4:101015.
Lasser SA, Ozbay Kurt FG, Arkhypov I, Utikal J, Umansky V. Myeloid-derived suppressor cells in cancer and cancer therapy. Nat Rev Clin Oncol. 2024;21:147–64.
Weber R, Groth C, Lasser S, Arkhypov I, Petrova V, Altevogt P, et al. IL-6 as a major regulator of MDSC activity and possible target for cancer immunotherapy. Cell Immunol. 2021;359:104254.
Neo SY, Tong L, Chong J, Liu Y, Jing X, Oliveira MMS, et al. Tumor-associated NK cells drive MDSC-mediated tumor immune tolerance through the IL-6/STAT3 axis. Sci Transl Med. 2024;16:eadi2952.
Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol. 2021;21:485–98.
Kumar A, Taghi Khani A, Sanchez Ortiz A, Swaminathan S. GM-CSF: A Double-Edged Sword in Cancer Immunotherapy. Front Immunol. 2022;13:901277.
Cordonnier M, Nardin C, Chanteloup G, Derangere V, Algros MP, Arnould L, et al. Tracking the evolution of circulating exosomal-PD-L1 to monitor melanoma patients. J Extracell Vesicles. 2020;9:1710899.
Ebrahimi A, Sevinc K, Gurhan Sevinc G, Cribbs AP, Philpott M, Uyulur F, et al. Bromodomain inhibition of the coactivators CBP/EP300 facilitate cellular reprogramming. Nat Chem Biol. 2019;15:519–28.
Raisner R, Kharbanda S, Jin L, Jeng E, Chan E, Merchant M, et al. Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep. 2018;24:1722–9.
Lu J, Luo Y, Rao D, Wang T, Lei Z, Chen X, et al. Myeloid-derived suppressor cells in cancer: therapeutic targets to overcome tumor immune evasion. Exp Hematol Oncol. 2024;13:39.
Li K, Shi H, Zhang B, Ou X, Ma Q, Chen Y, et al. Myeloid-derived suppressor cells as immunosuppressive regulators and therapeutic targets in cancer. Signal Transduct Target Ther. 2021;6:362.
Lechner MG, Liebertz DJ, Epstein AL. Characterization of cytokine-induced myeloid-derived suppressor cells from normal human peripheral blood mononuclear cells. J Immunol. 2010;185:2273–84.
Acknowledgements
This work was supported by NIH grants R01 CA256893 (XL), R01 CA264652 (XL), R01 CA157429 (XL), R01 CA272483 (XL). This research was supported by pilot funding (JL) provided by the Support from the University of Kentucky Markey Cancer Center’s Cancer Center Support Grant (P30 CA177558). The work was also supported by Biospecimen Procurement and Translational Pathology, Biostatistics and Bioinformatics, Flow Cytometry and Immune Monitoring Shared Resources of the University of Kentucky Markey Cancer Center (P30CA177558).
Author information
Authors and Affiliations
Contributions
XL designed and supervised the project. JL designed and performed the experiments, analyzed the data, and wrote the manuscript. XW conducted xenograft tumor harvesting and flow cytometry analysis. DH performed RNA-seq data analysis. HM and MN carried out the CRISPR knockout assays. MW and JP performed and evaluated immunofluorescence staining. XR, RW, SW, JW, and ZL contributed to manuscript editing. NB, NP, and KF developed and provided CCS1477.
Corresponding authors
Ethics declarations
Competing interests
NB, NP, and KF are employees and shareholders of CellCentric Ltd. Additionally, Neil Pegg serves as a board director and is listed as the inventor on patents related to CCS1477.
Ethics approval and consent to participate
All procedures involving animals were carried out in compliance with institutional guidelines and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Kentucky (Protocol No.: 2020-3685). No human subjects were involved in this study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, J., Wang, X., He, D. et al. Therapeutic targeting of the p300/CBP bromodomain enhances the efficacy of immune checkpoint blockade therapy. Oncogene 44, 2386–2395 (2025). https://doi.org/10.1038/s41388-025-03417-w
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03417-w
This article is cited by
-
CBP/p300, a promising therapeutic target for prostate cancer
Journal of Translational Medicine (2025)