Abstract
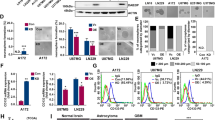
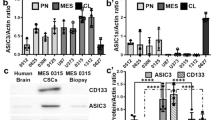
Glioblastoma (GBM) is an aggressive brain tumor driven by glioma stem cells (GSCs), which contribute to tumor growth and therapeutic resistance. This study investigates the effects of Gsk3β inhibition on GSC viability, focusing on the role of the canonical WNT signaling pathway. We found that Gsk3β inhibition activates the WNT pathway, leading to upregulation of Wwc1, which downregulates Yap via Lats1 phosphorylation. This reduces GSC proliferation, self-renewal, and enhances chemosensitivity. Analysis of clinical datasets revealed that WNT pathway activation correlates with improved prognosis in proneural gliomas, particularly in IDH1-mutated tumors. Our findings suggest that targeting the WNT-WWC1-YAP axis, particularly through Gsk3β inhibition, could induce synthetic lethality in GSCs and provide a promising therapeutic strategy for gliomas. These results highlight the potential of exploiting WNT-induced synthetic lethality as a novel approach for glioma treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Any additional information related to this manuscript is available from the lead contact upon request.
References
Ostrom QT, Price M, Neff C, Cioffi G, Waite KA, Kruchko C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2015–2019. Neuro Oncol. 2022;24:v1–v95. https://doi.org/10.1093/neuonc/noac202.
Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. https://doi.org/10.1016/j.ccr.2009.12.020.
Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor Evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2018;33:152 https://doi.org/10.1016/j.ccell.2017.06.003.
Xie XP, Laks DR, Sun D, Ganbold M, Wang Z, Pedraza AM, et al. Quiescent human glioblastoma cancer stem cells drive tumor initiation, expansion, and recurrence following chemotherapy. Dev Cell. 2022;57:32–46.e38. https://doi.org/10.1016/j.devcel.2021.12.007.
Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6. https://doi.org/10.1038/nature11287.
Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. https://doi.org/10.1038/nature03128.
Wu M, Wang T, Ji N, Lu T, Yuan R, Wu L, et al. Multi-omics and pharmacological characterization of patient-derived glioma cell lines. Nat Commun. 2024;15:6740. https://doi.org/10.1038/s41467-024-51214-y.
Shi Y, Lim SK, Liang Q, Iyer SV, Wang HY, Wang Z, et al. Gboxin is an oxidative phosphorylation inhibitor that targets glioblastoma. Nature. 2019;567:341–6. https://doi.org/10.1038/s41586-019-0993-x.
Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97–106. https://doi.org/10.1038/nrclinonc.2010.196.
Nusse R, Clevers H. Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–99. https://doi.org/10.1016/j.cell.2017.05.016.
Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. https://doi.org/10.1038/nrc3419.
Phoenix TN, Patmore DM, Boop S, Boulos N, Jacus MO, Patel YT, et al. Medulloblastoma genotype dictates blood brain barrier phenotype. Cancer Cell. 2016;29:508–22. https://doi.org/10.1016/j.ccell.2016.03.002.
Chang L, Jung NY, Atari A, Rodriguez DJ, Kesar D, Song TY, et al. Author Correction: Systematic profiling of conditional pathway activation identifies context-dependent synthetic lethalities. Nat Genet. 2024;56:553. https://doi.org/10.1038/s41588-024-01680-3.
Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. https://doi.org/10.1016/j.ccr.2008.12.006.
Uhrbom L, Hesselager G, Nistér M, Westermark B. Induction of brain tumors in mice using a recombinant platelet-derived growth factor B-chain retrovirus. Cancer Res. 1998;58:5275–9.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47 https://doi.org/10.1093/nar/gkv007.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. https://doi.org/10.1073/pnas.0506580102.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. https://doi.org/10.1093/bioinformatics/bts635.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. https://doi.org/10.1093/bioinformatics/btt656.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. https://doi.org/10.1093/bioinformatics/btp616.
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. https://doi.org/10.1186/1471-2105-14-7.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. https://doi.org/10.1093/bioinformatics/btr260.
Broos S, Hulpiau P, Galle J, Hooghe B, Van Roy F, De Bleser P. ConTra v2: a tool to identify transcription factor binding sites across species, update 2011. Nucleic Acids Res. 2011;39:W74–78. https://doi.org/10.1093/nar/gkr355.
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. https://doi.org/10.1038/nature07385.
Zhao Z, Meng F, Wang W, Wang Z, Zhang C, Jiang T. Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Sci Data. 2017;4:170024. https://doi.org/10.1038/sdata.2017.24.
Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. https://doi.org/10.1016/j.ccr.2010.03.017.
Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–46. https://doi.org/10.1016/j.ccr.2013.08.001.
Zhong Z, Jiao Z, Yu FX. The Hippo signaling pathway in development and regeneration. Cell Rep. 2024;43:113926 https://doi.org/10.1016/j.celrep.2024.113926.
Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–16. https://doi.org/10.1016/j.devcel.2009.12.013.
Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–99. https://doi.org/10.1016/j.devcel.2009.12.012.
Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–96. https://doi.org/10.1007/s00401-011-0800-8.
Northcott PA, Robinson GW, Kratz CP, Mabbott DJ, Pomeroy SL, Clifford SC, et al. Medulloblastoma. Nat Rev Dis Prim. 2019;5:11 https://doi.org/10.1038/s41572-019-0063-6.
Fernandez LA, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–41. https://doi.org/10.1101/gad.1824509.
Ren F, Shi Q, Chen Y, Jiang A, Ip YT, Jiang H, et al. Drosophila Myc integrates multiple signaling pathways to regulate intestinal stem cell proliferation during midgut regeneration. Cell Res. 2013;23:1133–46. https://doi.org/10.1038/cr.2013.101.
Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–97. https://doi.org/10.1016/j.cell.2014.06.003.
Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–84. https://doi.org/10.1016/j.cell.2014.06.004.
Atkinson JM, Rank KB, Zeng Y, Capen A, Yadav V, Manro JR, et al. Activating the Wnt/β-catenin pathway for the treatment of melanoma-application of LY2090314, a novel selective inhibitor of glycogen synthase kinase-3. PLoS ONE. 2015;10:e0125028 https://doi.org/10.1371/journal.pone.0125028.
Korur S, Huber RM, Sivasankaran B, Petrich M, Morin P Jr., Hemmings BA, et al. GSK3beta regulates differentiation and growth arrest in glioblastoma. PLoS ONE. 2009;4:e7443 https://doi.org/10.1371/journal.pone.0007443.
Kageshita T, Hamby CV, Ishihara T, Matsumoto K, Saida T, Ono T. Loss of beta-catenin expression associated with disease progression in malignant melanoma. Br J Dermatol. 2001;145:210–6. https://doi.org/10.1046/j.1365-2133.2001.04336.x.
Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, et al. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131:2791–801. https://doi.org/10.1242/dev.01165.
Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–34. https://doi.org/10.1101/gad.1726608.
Panciera T, Azzolin L, Fujimura A, Di Biagio D, Frasson C, Bresolin S, et al. Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell. 2016;19:725–37. https://doi.org/10.1016/j.stem.2016.08.009.
Hansen CG, Moroishi T, Guan KL. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 2015;25:499–513. https://doi.org/10.1016/j.tcb.2015.05.002.
Guske K, Schmitz B, Schelleckes M, Duning K, Kremerskothen J, Pavenstädt HJ, et al. Tissue-specific differences in the regulation of KIBRA gene expression involve transcription factor TCF7L2 and a complex alternative promoter system. J Mol Med. 2014;92:185–96. https://doi.org/10.1007/s00109-013-1089-y.
Thorne CA, Wichaidit C, Coster AD, Posner BA, Wu LF, Altschuler SJ. GSK-3 modulates cellular responses to a broad spectrum of kinase inhibitors. Nat Chem Biol. 2015;11:58–63. https://doi.org/10.1038/nchembio.1690.
Acknowledgements
The authors thank Dr. Wei Mo for HTS cells and Dr. Shangfeng Gao for GBM paraffin sections. This work was funded by the National Key Research and Development Program of China (#2022YFA1103900 to JC), the Changping Laboratory and the CAMS Innovation Fund for Medical Sciences (CIFMS) (#2024-I2M-3-022 to JC). Additional funds to JC are from the Chinese Institute for Brain Research and Changping Laboratory.
Author information
Authors and Affiliations
Contributions
FFR, XZL, TL, YLY, GC and SH performed the experiments. JC designed the experiments. FFR, LFP and JC analyzed the data and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All the experimental procedures were performed in compliance with the protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Soochow University or the Chinese Institute for Brain Research. All mouse experiments were approved by and performed according to the institutional guidelines.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ren, F., Yi, Y., Lu, T. et al. Synthetic lethality through Gsk3β inhibition in glioma stem cells via the WNT-WWC1-YAP axis. Oncogene 44, 2427–2439 (2025). https://doi.org/10.1038/s41388-025-03418-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03418-9
This article is cited by
-
Immunometabolism: crosstalk with tumor metabolism and implications for cancer immunotherapy
Molecular Cancer (2025)