Abstract
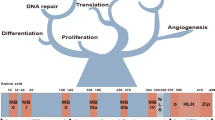
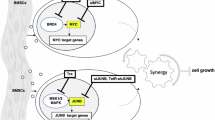
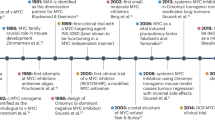
The MYC oncogene is a frequently activated oncogene in human cancers, and its high expression is strongly correlated with a poor prognosis. The lack of conventional enzyme-binding sites in MYC poses significant challenges for the development of small-molecule-based therapies to treat MYC-deregulated cancer. In particular, only one transmembrane peptide that targets c-MYC has advanced to early clinical trials, thus highlighting the need of effective and direct approaches for targeting c-MYC in cancer treatment. In this study, we developed a conjugated nanobody (NB) that specifically targets MYC, termed a cell-permeable MYC-targeting nanobody (CPMycNB), via sortase-mediated protein ligation. CPMycNB effectively entered the nucleus and bound to c-MYC, thereby disrupting the c-MYC-MAX interaction. This disruption resulted in the downregulation of c-MYC-targeted genes, activation of apoptotic pathways, and inhibition of cell growth and proliferation in c-MYC-driven tumor cells. Using hydrogen-deuterium exchange mass spectrometry, we found that CPMycNB interacted with the leucine zipper domain of c-MYC. Furthermore, xenograft studies confirmed the therapeutic efficacy of CPMycNB, which significantly reduced tumor size and weight. Our findings highlight the potential of CPMycNB for the treatment of c-MYC-associated malignancies.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Dang CV. Myc on the path to cancer. Cell. 2012;149:22–35.
Schaub FX, Dhankani V, Berger AC, Trivedi M, Richardson AB, Shaw R, et al. Pan-cancer alterations of the myc oncogene and its proximal network across the cancer genome atlas. Cell Syst. 2018;6:282–300.
Wasylishen AR, Penn LZ. Myc: the beauty and the beast. Genes cancer. 2010;1:532–41.
Lin CY, Lovn J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-myc. Cell. 2012;151:56–67.
Di Giacomo S, Sollazzo M, Paglia S, Grifoni D. Myc, cell competition, and cell death in cancer: the inseparable triad. Genes. 2017;8:120.
Nau MM, Brooks BJ, Battey J, Sausville E, Gazdar AF, Kirsch IR, et al. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985;318:69–73.
Zhou ZQ, Hurlin PJ. The interplay between mad and myc in proliferation and differentiation. Trends cell Biol. 2001;11:S10–S14.
Bretones G, Delgado MD, Len J. Myc and cell cycle control. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2015;1849:506–16.
Llombart V, Mansour MR. Therapeutic targeting of undruggable MYC. EBioMedicine. 2022;75:103756.
Gustafson W, Weiss W. Myc proteins as therapeutic targets. Oncogene. 2010;29:1249–59.
Mo H, Henriksson M. Identification of small molecules that induce apoptosis in a myc-dependent manner and inhibit myc-driven transformation. Proc Natl Acad Sci. 2006;103:6344–9.
Duffy MJ, O’Grady S, Tang M, Crown J. Myc as a target for cancer treatment. Cancer Treat Rev. 2021;94:102154.
Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. Bet bromodomain inhibition as a therapeutic strategy to target c-myc. Cell. 2011;146:904–17.
Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AMK, et al. Protac-induced bet protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci. 2016;113:7124–9.
Crump NT, Ballabio E, Godfrey L, Thorne R, Repapi E, Kerry J, et al. Bet inhibition disrupts transcription but retains enhancer-promoter contact. Nat Commun. 2021;12:223.
Han H, Jain AD, Truica MI, Izquierdo-Ferrer J, Anker JF, Lysy B, et al. Small-molecule myc inhibitors suppress tumor growth and enhance immunotherapy. Cancer cell. 2019;36:483–97.
Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, et al. Small-molecule antagonists of myc/max dimerization inhibit myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci. 2002;99:3830–5.
Amati B. Myc degradation: dancing with ubiquitin ligases. Proc Natl Acad Sci. 2004;101:8843–4.
Crawford LJ, Campbell DC, Morgan JJ, Lawson MA, Down JM, Chauhan D, et al. The e3 ligase huwe1 inhibition as a therapeutic strategy to target myc in multiple myeloma. Oncogene. 2020;39:5001–14.
Ross J, Miron CE, Plescia J, Laplante P, McBride K, Moitessier N, et al. Targeting myc: from understanding its biology to drug discovery. Eur J Med Chem. 2021;213:113137.
Donati G, Amati B. Myc and therapy resistance in cancer: risks and opportunities. Mol Oncol. 2022;16:3828–54.
Wang E, Sorolla A, Cunningham PT, Bogdawa HM, Beck S, Golden E, et al. Tumor penetrating peptides inhibiting myc as a potent targeted therapeutic strategy for triple-negative breast cancers. Oncogene. 2019;38:140–50.
Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, et al. Modelling myc inhibition as a cancer therapy. Nature. 2008;455:679–83.
Demma MJ, Mapelli C, Sun A, Bodea S, Ruprecht B, Javaid S. et al. Omomyc reveals new mechanisms to inhibit the myc oncogene. Mol Cellular Biol. 2019;39:00248–19.
Beaulieu ME, Jauset T, Mass-Valls D, Martnez-Martn S, Rahl P, Maltais L, et al. Intrinsic cell-penetrating activity propels omomyc from proof of concept to viable anti-myc therapy. Sci Transl Med. 2019;11:eaar5012.
Garralda E, Beaulieu ME, Moreno V, Casacuberta-Serra S, Martnez-Martn S, Foradada L, et al. Myc targeting by omo-103 in solid tumors: a phase 1 trial. Nat Med. 2024;30:762–71.
Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–97.
Muyldermans S, Atarhouch T, Saldanha J, Barbosa J, Hamers R. Sequence and structure of vh domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng Des Selection. 1994;7:1129–35.
Ji, F, Ren, J, Vincke, C, Jia, L, Muyldermans, S Nanobodies: From serendipitous discovery of heavy chain-only antibodies in camelids to a wide range of useful applications. In: Single-Domain Antibodies: Methods and Protocols. Springer. 2022:3-17.
Daley-Bauer L, Purdy S, Smith M, Gagliardo L, Davis W, Appleton J. Contributions of conventional and heavy-chain igg to immunity in fetal, neonatal, and adult alpacas. Clin Vaccin Immunol. 2010;17:2007–15.
Conrath K, Wernery U, Muyldermans S, Nguyen VK. Emergence and evolution of functional heavy-chain antibodies in camelidae. Dev Comp Immunol. 2003;27:87–103.
Atibalentja DF, Deutzmann A, Felsher DW. A big step for myc-targeted therapies. Trends Cancer. 2024;10:383–85.
Pooga M, Langel Ü. Classes of Cell-Penetrating Peptides. Methods Mol Biol. 2015;1324:3–28.
Raucher D, Ryu JS. Cell-penetrating peptides: strategies for anticancer treatment. Trends Mol Med. 2015;21:560–70.
Wolfe JM, Fadzen CM, Choo ZN, Holden RL, Yao M, Hanson GJ, et al. Machine learning to predict cell-penetrating peptides for antisense delivery. ACS Cent Sci. 2018;4:512–20.
Kadonosono T, Yamano A, Goto T. et al. Cell penetrating peptides improve tumor delivery of cargos through neuropilin-1-dependent extravasation. J Control Release. 2015;201:14–21.
Yu S, Yang H, Li T, Pan H, Ren S, Luo G, et al. Efficient intracellular delivery of proteins by a multifunctional chimaeric peptide in vitro and in vivo. Nat Commun. 2021;12:5131.
Teo SL, Rennick JJ, Yuen D, Al-Wassiti H, Johnston AP, Pouton CW. Unravelling cytosolic delivery of cell penetrating peptides with a quantitative endosomal escape assay. Nat Commun. 2021;12:3721.
Kent, L Targeting then-myc oncoprotein using nanobody technology. Ph.D. thesis; University of Cambridge. 2018.
Mundo L, Ambrosio MR, Raimondi F, Del Porro L, Guazzo R, Mancini V, et al. Molecular switch from myc to mycn expression in myc protein negative burkitt lymphoma cases. Blood Cancer J. 2019;9:91.
Wang C, Zhang J, Yin J, Gan Y, Xu S, Gu Y, et al. Alternative approaches to target myc for cancer treatment. Signal Transduct Target Ther. 2021;6:117.
Ingvarsson S. The myc gene family proteins and their role in transformation and differentiation. Semin Cancer Biol. 1990;1:359–69.
Theile CS, Witte MD, Blom AE, Kundrat L, Ploegh HL, Guimaraes CP. Site-specific n-terminal labeling of proteins using sortase-mediated reactions. Nat Protoc. 2013;8:1800–7.
Massa S, Vikani N, Betti C, Ballet S, Vanderhaegen S, Steyaert J, et al. Sortase a-mediated site-specific labeling of camelid single-domain antibody-fragments: a versatile strategy for multiple molecular imaging modalities. Contrast media Mol imaging. 2016;11:328–39.
Zhu WL, Shin SY. Effects of dimerization of the cell-penetrating peptide tat analog on antimicrobial activity and mechanism of bactericidal action. J Pept Sci: Publ Eur Pept Soc. 2009;15:345–52.
Brooks H, Lebleu B, Vivs E. Tat peptide-mediated cellular delivery: back to basics. Adv drug Deliv Rev. 2005;57:559–77.
Zhang H, Zhang Y, Zhang C, Yu H, Ma Y, Li Z, et al. Recent advances of cell-penetrating peptides and their application as vectors for delivery of peptide and protein-based cargo molecules. Pharmaceutics. 2023;15:2093.
Rosenblum D, Joshi N, Tao W, Karp JM, Peer D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat Commun. 2018;9:1410.
Hsiue EHC, Wright KM, Douglass J, Hwang MS, Mog BJ, Pearlman AH, et al. Targeting a neoantigen derived from a common tp53 mutation. Science. 2021;371:eabc8697.
Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74.
Pearlman AH, Hwang MS, Konig MF, Hsiue EHC, Douglass J, DiNapoli SR, et al. Targeting public neoantigens for cancer immunotherapy. Nat cancer. 2021;2:487–97.
Acknowledgements
This study was supported by the National Key Research and Development Program of China (Grant No. 2021YFA1302601), the National Natural Science Foundation of China (Grant No. 82172556), the Beijing Natural Science Foundation (IS24040), and the National Natural Science Foundation of China (Grant No. T2293763). We greatly appreciate the Facility for Protein Chemistry and Proteomics at Tsinghua University for sample analysis.
Author information
Authors and Affiliations
Contributions
YX conceived and coordinated the entire project, designed the experiments, conducted data analysis, and authored the manuscript. HJ, TL, XT, and ZZ performed the experiments and analyzed the data. WD and YW contributed to data analysis and the interpretation of results. ZM and HD were responsible for additional data analysis and critically reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xue, Y., Jiang, H., Li, T. et al. Inhibition of tumor growth using A conjugated nanobody that specifically targets c-MYC. Oncogene 44, 3213–3224 (2025). https://doi.org/10.1038/s41388-025-03486-x
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03486-x
This article is cited by
-
MYC as a Target for Cancer Treatment: from Undruggable to Druggable?
Targeted Oncology (2025)