Abstract
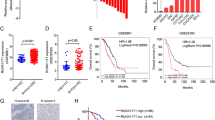
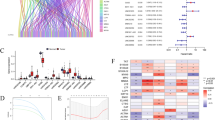
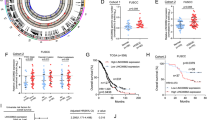
Ovarian cancer (OC) remains a significant challenge in oncology due to its late diagnosis and poor prognosis. Emerging evidence suggests that long non-coding RNAs (lncRNAs) play critical roles in cancer biology. Herein, we reported that LINC00654 was highly expressed in OC tissues and correlated with poor patient prognosis. In addition, LINC00654 silencing restrained OC cell proliferation and migration in vitro and in vivo. Mechanically, LINC00654 was identified to directly interact with Human antigen R (HuR), a known RNA-binding protein, through RNA pull-down, RNA immunoprecipitation (RIP), and cross‑linking immunoprecipitation (CLIP). Further analysis revealed that LINC00654 could induce the translocation of HuR from the nucleus to the cytosol, where it regulated the stability of its target oncogenes, such as VASH2. The stabilization of VASH2 subsequently activated the TGF-β pathway, which is known to play a critical role in cancer progression. Taken together, these findings establish a specific mechanism by which LINC00654 interacts with HuR, facilitates its nuclear export, and stabilizes VASH2, thereby activating the TGF-β pathway and promoting OC progression. This insight into LINC00654’s role in OC provides potential therapeutic targets for intervention.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
The mass spectrometry proteomics data are available in the ProteomeXchange Consortium through the PRIDE partner repository (accession number PXD051820). The RNA-Seq datasets generated during the current study are available in the ArrayExpress repository (accession number E-MTAB-14741). All data are available from the corresponding author upon reasonable request.
References
Kuroki L, Guntupalli SR. Treatment of epithelial ovarian cancer. BMJ. 2020;371:m3773.
Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3–14.
Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393:1240–53.
Sundar S, Neal RD, Kehoe S. Diagnosis of ovarian cancer. BMJ. 2015;351:h4443.
Konstantinopoulos PA, Matulonis UA. Clinical and translational advances in ovarian cancer therapy. Nat Cancer. 2023;4:1239–57.
Tan Y-T, Lin J-F, Li T, Li J-J, Xu R-H, Ju H-Q. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun. 2021;41:109–20.
Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–81.
Zhan L, Li J, Wei B. Long non-coding RNAs in ovarian cancer. J Exp Clin Cancer Res. 2018;37:120.
Xie W, Sun H, Li X, Lin F, Wang Z, Wang X. Ovarian cancer: epigenetics, drug resistance, and progression. Cancer Cell Int. 2021;21:434.
Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62.
Herman AB, Tsitsipatis D, Gorospe M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell. 2022;82:2252–66.
Xu W, Zhou G, Wang H, Liu Y, Chen B, Chen W, et al. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer. 2020;146:2901–12.
Chen Y, Li C, Wang N, Wu Z, Zhang J, Yan J, et al. Identification of LINC00654-NINL regulatory axis in diffuse large B-cell lymphoma in silico analysis. Front Oncol. 2022;12:883301.
Li L, Gan ZH, Qin L, Jiao SH, Shi Y. AIB1 regulates the ovarian cancer cell cycle through TUG1. Eur Rev Med Pharmacol Sci. 2017;21:5610–7.
de Silanes López, Zhan I, Lal M, Yang A, Gorospe X. M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–92.
Gorospe M. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle. 2003;2:412–4.
Simion V, Zhou H, Haemmig S, Pierce JB, Mendes S, Tesmenitsky Y, et al. A macrophage-specific lncRNA regulates apoptosis and atherosclerosis by tethering HuR in the nucleus. Nat Commun. 2020;11:6135.
Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA. 2010;1:214–29.
Lal S, Burkhart RA, Beeharry N, Bhattacharjee V, Londin ER, Cozzitorto JA, et al. HuR posttranscriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells. Cancer Res. 2014;74:1128–40.
Zhou J, Li J, Qian C, Qiu F, Shen Q, Tong R, et al. LINC00624/TEX10/NF-κB axis promotes proliferation and migration of human prostate cancer cells. Biochem Biophys Res Commun. 2022;601:1–8.
Yan Y, Tao H, He J, Huang S-Y. The HDOCK server for integrated protein-protein docking. Nat Protoc. 2020;15:1829–52.
Weng G, Wang E, Wang Z, Liu H, Zhu F, Li D, et al. HawkDock: a web server to predict and analyze the protein-protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019;47:W322–W30.
Zhou Q, Guo Y, Zheng B, Shao B, Jiang M, Wang G, et al. Establishment of a proteome profile and identification of molecular markers for mouse spermatogonial stem cells. J Cell Mol Med. 2015;19:521–34.
Zhang K, Xu J, Ding Y, Shen C, Lin M, Dai X, et al. BMI1 promotes spermatogonia proliferation through epigenetic repression of Ptprm. Biochem Biophys Res Commun. 2021;583:169–77.
Wang Q, Wu Y, Lin M, Wang G, Liu J, Xie M, et al. BMI1 promotes osteosarcoma proliferation and metastasis by repressing the transcription of SIK1. Cancer Cell Int. 2022;22:136.
Wang M, Chen X, Wu Y, Zheng Q, Chen W, Yan Y, et al. RpS13 controls the homeostasis of germline stem cell niche through Rho1-mediated signals in the Drosophila testis. Cell Prolif. 2020;53:e12899.
Liu Y, Yu X, Huang A, Zhang X, Wang Y, Geng W, et al. INTS7-ABCD3 interaction stimulates the proliferation and osteoblastic differentiation of mouse bone marrow mesenchymal stem cells by suppressing oxidative stress. Front Physiol. 2021;12:758607.
Luo X, Huang Y, Li H, Luo Y, Zuo Z, Ren J, et al. SPENCER: a comprehensive database for small peptides encoded by noncoding RNAs in cancer patients. Nucleic Acids Res. 2022;50:D1373–D81.
Gruber AR, Fallmann J, Kratochvill F, Kovarik P, Hofacker IL. AREsite: a database for the comprehensive investigation of AU-rich elements. Nucleic Acids Res. 2011;39:D66–D9.
Brennan CM. Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–77.
Long J, Zhao W, Xiang Y, Wang Y, Xiang W, Liu X, et al. STAT3 promotes cytoplasmic-nuclear translocation of RNA-binding protein HuR to inhibit IL-1β-induced IL-8 production. Int Immunopharmacol. 2024;133:112065.
Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8.
Kulkarni AA, Kingsbury SR, Tudzarova S, Hong H-K, Loddo M, Rashid M, et al. Cdc7 kinase is a predictor of survival and a novel therapeutic target in epithelial ovarian carcinoma. Clin Cancer Res. 2009;15:2417–25.
Han X, Fang Z, Wang H, Jiao R, Zhou J, Fang N. CUL4A functions as an oncogene in ovarian cancer and is directly regulated by miR-494. Biochem Biophys Res Commun. 2016;480:675–81.
Wu J, Wei J-J. HMGA2 and high-grade serous ovarian carcinoma. J Mol Med. 2013;91:1155–65.
Zeng Z, Ji N, Yi J, Lv J, Yuan J, Lin Z, et al. LGR4 overexpression is associated with clinical parameters and poor prognosis of serous ovarian cancer. Cancer Biomark. 2020;28:65–72.
Feng C, Han Y-H, Qi N, Li J, Sheng Q-H, Liu Y, et al. Functional implications of PABPC1 in the development of ovarian cancer. Open Med. 2021;16:805–15.
Norita R, Suzuki Y, Furutani Y, Takahashi K, Yoshimatsu Y, Podyma-Inoue KA, et al. Vasohibin-2 is required for epithelial-mesenchymal transition of ovarian cancer cells by modulating transforming growth factor-β signaling. Cancer Sci. 2017;108:419–26.
Tu M, Li Z, Liu X, Lv N, Xi C, Lu Z, et al. Vasohibin 2 promotes epithelial-mesenchymal transition in human breast cancer via activation of transforming growth factor β 1 and hypoxia dependent repression of GATA-binding factor 3. Cancer Lett. 2017;388:187–97.
Chia Z-J, Cao Y-N, Little PJ, Kamato D. Transforming growth factor-β receptors: versatile mechanisms of ligand activation. Acta Pharm Sin. 2024;45:1337–48.
Gobbo F, Martelli F, Di Virgilio A, Demaria E, Sarli G, Migliaccio AR. The variation in the traits ameliorated by inhibitors of JAK1/2, TGF-β, P-selectin, and CXCR1/CXCR2 in the Gata1low model suggests that myelofibrosis should be treated by these drugs in combination. Int J Mol Sci. 2024;25:7703.
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8.
Xie M, Ma T, Xue J, Ma H, Sun M, Zhang Z, et al. The long intergenic non-protein coding RNA 707 promotes proliferation and metastasis of gastric cancer by interacting with mRNA stabilizing protein HuR. Cancer Lett. 2019;443:67–79.
Du Y, Xu X, Lv S, Liu H, Sun H, Wu J. SOCS7/HuR/FOXM1 signaling axis inhibited high-grade serous ovarian carcinoma progression. J Exp Clin Cancer Res. 2022;41:185.
Erkinheimo T-L, Lassus H, Sivula A, Sengupta S, Furneaux H, Hla T, et al. Cytoplasmic HuR expression correlates with poor outcome and with cyclooxygenase 2 expression in serous ovarian carcinoma. Cancer Res. 2003;63:7591–4.
Zhang M, Xu Y, Zhang Y, Li B, Lou G. Circular RNA circE2F2 promotes malignant progression of ovarian cancer cells by upregulating the expression of E2F2 protein via binding to HuR protein. Cell Signal. 2021;84:110014.
Schultz CW, Preet R, Dhir T, Dixon DA, Brody JR. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR). Wiley Interdiscip Rev RNA. 2020;11:e1581.
Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA. 2017;8. https://doi.org/10.1002/wrna.1372.
Finan JM, Sutton TL, Dixon DA, Brody JR. Targeting the RNA-binding protein HuR in cancer. Cancer Res. 2023;83:3507–16.
Du H, Zhao J, Hai L, Wu J, Yi H, Shi Y. The roles of vasohibin and its family members: beyond angiogenesis modulators. Cancer Biol Ther. 2017;18:827–32.
Bowler E, Oltean S. Alternative splicing in angiogenesis. Int J Mol Sci. 2019;20:2067.
Liu S, Ren J, Ten Dijke P. Targeting TGFβ signal transduction for cancer therapy. Signal Transduct Target Ther. 2021;6:8.
Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50:924–40.
Yeung T-L, Leung CS, Wong K-K, Samimi G, Thompson MS, Liu J, et al. TGF-β modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 2013;73:5016–28.
Kumari A, Shonibare Z, Monavarian M, Arend RC, Lee NY, Inman GJ, et al. TGFβ signaling networks in ovarian cancer progression and plasticity. Clin Exp Metastasis. 2021;38:139–61.
Alsina-Sanchis E, Figueras A, Lahiguera Á, Vidal A, Casanovas O, Graupera M, et al. The TGFβ pathway stimulates ovarian cancer cell proliferation by increasing IGF1R levels. Int J Cancer. 2016;139:1894–903.
Funding
This work was supported by the National Natural Science Foundation of China (82001576) and the Suzhou Gu Su Health Talent Research Project (GSWS2023056).
Author information
Authors and Affiliations
Contributions
CS: conceptualization, funding acquisition, investigation, supervision, writing revised draft. RC: data curation, formal analysis, writing – original draft. QZ: investigation, data curation. TZ: data curation, visualization. TTW, investigation, data curation. WG, investigation, methodology. GW: investigation, validation. GF: conceptualization, supervision, project administration. LQ: funding acquisition, supervision. TW: supervision, project administration.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
All human procedures received approval from the Ethical Committee of the Affiliated Suzhou Hospital of Nanjing Medical University (No. 2022012), adhering to the World Medical Association’s Declaration of Helsinki, and written informed consent was obtained from each patient. All animal procedures were approved by the Animal Ethics Committee of Nanjing Medical University (No. 2402015) and performed according to the ARRIVE guidelines.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shen, C., Cao, R., Zhou, Q. et al. LINC00654 promotes ovarian cancer progression by facilitating nuclear export of HuR and stabilizing oncogenic mRNAs. Oncogene 44, 3422–3436 (2025). https://doi.org/10.1038/s41388-025-03500-2
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03500-2