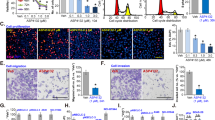
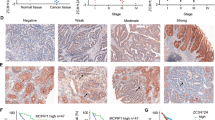
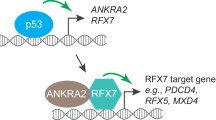
Abstract
Activated Cdc42-associated kinase 1 (ACK1) is an oncogenic non-receptor kinase that promotes tumor cell survival and impairs T-cell activation. Targeting ACK1 has great promise in cancer control. However, tumor adaptive responses that may limit the anticancer efficacy of ACK1 inhibition (ACK1i) remain unclear. We found that ACK1i treatment triggered the PINK1/PARKIN-mediated adaptive mitophagy by upregulating the mitophagy receptor BNIP3. Mass/Spectrometry and co-immunoprecipitation (Co-IP) results indicated that ACK1 interacted with transcription factor regulatory factor X 2 (RFX2) through its MHR domain, and competitively inhibits RFX2 ubiquitination via the E3 ubiquitin ligase MIB1. Conversely, ACK1i facilitates MIB1-mediated RFX2 ubiquitination and degradation. Moreover, we observed that RFX2 is a transcriptional suppressor of BNIP3 using luciferase reporter gene assays and chromatin immunoprecipitation (ChIP). Overall, ACK1i treatment causes RFX2 instability and thereby diminishes RFX2’s suppressive effects on BNIP3 transcription, leading to BNIP3 accumulation and the activation of mitophagy pathways. This adaptive mitophagy allows NSCLC cells to survive under ACK1 inhibition, potentially reducing the efficacy of ACK1i. ACK1i combined with mitophagy-inhibiting agents may attain a more accomplished response in NSCLC. In conclusion, ACK1i induced mitophagy through the release of RFX2 inhibition on BNIP3 transcription, thereby driving adaptive resistance. Inhibiting mitophagy sensitizes NSCLC to ACK1i.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Public data used in this work can be acquired from (TCGA, https://www.cancer.gov/tcga/). The raw experimental data and analysis codes supporting the conclusions of this article will be made available by the corresponding author.
References
Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17:637–58.
Cooper AJ, Sequist LV, Lin JJ. Third-generation EGFR and ALK inhibitors: mechanisms of resistance and management. Nat Rev Clin Oncol. 2022;19:499–514.
Mahajan K, Mahajan NP. ACK1/TNK2 tyrosine kinase: molecular signaling and evolving role in cancers. Oncogene. 2015;34:4162–7.
van der Horst EH, Degenhardt YY, Strelow A, Slavin A, Chinn L, Orf J, et al. Metastatic properties and genomic amplification of the tyrosine kinase gene ACK1. Proc Natl Acad Sci USA. 2005;102:15901–6.
Zhu J, Liu Y, Zhao M, Cao K, Ma J, Peng S. Identification of downstream signaling cascades of ACK1 and prognostic classifiers in non-small cell lung cancer. Aging. 2021;13:4482–502.
Hu F, Liu H, Xie X, Mei J, Wang M. Activated cdc42-associated kinase is up-regulated in non-small-cell lung cancer and necessary for FGFR-mediated AKT activation. Mol Carcinog. 2016;55:853–63.
Mendez P, Ramirez JL. Copy number gains of FGFR1 and 3q chromosome in squamous cell carcinoma of the lung. Transl Lung Cancer Res. 2013;2:101–11.
Chua BT, Lim SJ, Tham SC, Poh WJ, Ullrich A. Somatic mutation in the ACK1 ubiquitin association domain enhances oncogenic signaling through EGFR regulation in renal cancer derived cells. Mol Oncol. 2010;4:323–34.
Mahendrarajah N, Borisova ME, Reichardt S, Godmann M, Sellmer A, Mahboobi S, et al. HSP90 is necessary for the ACK1-dependent phosphorylation of STAT1 and STAT3. Cell Signal. 2017;39:9–17.
Kiweler N, Brill B, Wirth M, Breuksch I, Laguna T, Dietrich C, et al. The histone deacetylases HDAC1 and HDAC2 are required for the growth and survival of renal carcinoma cells. Arch Toxicol. 2018;92:2227–43.
Mahajan K, Coppola D, Challa S, Fang B, Chen YA, Zhu W, et al. Ack1 mediated AKT/PKB tyrosine 176 phosphorylation regulates its activation. PLoS One. 2010;5:e9646.
Mahajan NP, Whang YE, Mohler JL, Earp HS. Activated tyrosine kinase Ack1 promotes prostate tumorigenesis: role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer Res. 2005;65:10514–23.
Mahajan NP, Liu Y, Majumder S, Warren MR, Parker CE, Mohler JL, et al. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proc Natl Acad Sci USA. 2007;104:8438–43.
Nonami A, Sattler M, Weisberg E, Liu Q, Zhang J, Patricelli MP, et al. Identification of novel therapeutic targets in acute leukemias with NRAS mutations using a pharmacologic approach. Blood. 2015;125:3133–43.
Mahendrarajah N, Paulus R, Kramer OH. Histone deacetylase inhibitors induce proteolysis of activated CDC42-associated kinase-1 in leukemic cells. J Cancer Res Clin Oncol. 2016;142:2263–73.
Yokoyama N, Lougheed J, Miller WT. Phosphorylation of WASP by the Cdc42-associated kinase ACK1: dual hydroxyamino acid specificity in a tyrosine kinase. J Biol Chem. 2005;280:42219–26.
Mahajan K, Malla P, Lawrence HR, Chen Z, Kumar-Sinha C, Malik R, et al. ACK1/TNK2 regulates histone H4 Tyr88-phosphorylation and AR gene expression in castration-resistant prostate cancer. Cancer Cell. 2017;31:790–803.e8.
Chouhan S, Sridaran D, Weimholt C, Luo J, Li T, Hodgson MC, et al. SHP2 as a primordial epigenetic enzyme expunges histone H3 pTyr-54 to amend androgen receptor homeostasis. Nat Commun. 2024;15:5629.
Chouhan S, Sawant M, Weimholt C, Luo J, Sprung RW, Terrado M, et al. TNK2/ACK1-mediated phosphorylation of ATP5F1A (ATP synthase F1 subunit alpha) selectively augments survival of prostate cancer while engendering mitochondrial vulnerability. Autophagy. 2023;19:1000–25.
Hu C, Zheng Z, Pang S, Zhu Y, Jie J, Lai Z, et al. Chimeric SFT2D2-TBX19 promotes prostate cancer progression by encoding TBX19-202 protein and stabilizing mitochondrial ATP synthase through ATP5F1A phosphorylation. Adv Sci. 2024;11:e2408426.
Li Q, Zhang T, Song P, Tong L, Feng F, Guo J, et al. Design, synthesis, and evaluation of (R)-8-((Tetrahydrofuran-2-yl)methyl)pyrido[2,3-d]pyrimidin-7-ones as novel selective ACK1 inhibitors to combat acquired resistance to the third-generation EGFR inhibitor. J Med Chem. 2023;66:6905–21.
Wang A, Pei J, Shuai W, Lin C, Feng L, Wang Y, et al. Small molecules targeting activated Cdc42-associated kinase 1 (ACK1/TNK2) for the treatment of cancers. J Med Chem. 2021;64:16328–48.
Kumar V, Kumar R, Parate S, Danishuddin, Lee G, Kwon M, et al. Identification of activated Cdc42-associated kinase inhibitors as potential anticancer agents using pharmacoinformatic approaches. Biomolecules. 2023;13:217.
Lawrence HR, Mahajan K, Luo Y, Zhang D, Tindall N, Huseyin M, et al. Development of novel ACK1/TNK2 inhibitors using a fragment-based approach. J Med Chem. 2015;58:2746–63.
Phatak SS, Zhang S. A novel multi-modal drug repurposing approach for identification of potent ACK1 inhibitors. Pac Symp Biocomput. 2013;29–40.
Sridaran D, Mahajan NP. ACK1/TNK2 kinase: molecular mechanisms and emerging cancer therapeutics. Trends Pharm Sci. 2025;46:62–77.
Saygin C, Giordano G, Shimamoto K, Eisfelder B, Thomas-Toth A, Venkataraman G, et al. Dual targeting of apoptotic and signaling pathways in T-lineage acute lymphoblastic leukemia. Clin Cancer Res. 2023;29:3151–61.
Zhang T, Qu R, Chan S, Lai M, Tong L, Feng F, et al. Discovery of a novel third-generation EGFR inhibitor and identification of a potential combination strategy to overcome resistance. Mol Cancer. 2020;19:90.
Pei J, Wang G, Wang A, Wu C, Pan X, Shuai W, et al. Design, synthesis, and antitumor activity of potent and selective EGFR L858R/T790M inhibitors and identification of a combination therapy to overcome acquired resistance in models of non-small-cell lung cancer. J Med Chem. 2023;66:5719–52.
Gu J, Qian L, Zhang G, Mahajan NP, Owonikoko TK, Ramalingam SS, et al. Inhibition of ACK1 delays and overcomes acquired resistance of EGFR mutant NSCLC cells to the third generation EGFR inhibitor, osimertinib. Lung Cancer. 2020;150:26–35.
Sridaran D, Chouhan S, Mahajan K, Renganathan A, Weimholt C, Bhagwat S, et al. Inhibiting ACK1-mediated phosphorylation of C-terminal Src kinase counteracts prostate cancer immune checkpoint blockade resistance. Nat Commun. 2022;13:6929.
Oh H, Hwang I, Jang JY, Wu L, Cao D, Yao J, et al. Therapy-Induced Transdifferentiation Promotes Glioma Growth Independent of EGFR Signaling. Cancer Res. 2021;81:1528–39.
Bonsignore G, Martinotti S, Ranzato E. Endoplasmic reticulum stress and cancer: could unfolded protein response be a druggable target for cancer therapy? Int J Mol Sci. 2023;24:1566.
Sniegocka M, Liccardo F, Fazi F, Masciarelli S. Understanding ER homeostasis and the UPR to enhance treatment efficacy of acute myeloid leukemia. Drug Resist Updat. 2022;64:100853.
Chern YJ, Tai IT. Adaptive response of resistant cancer cells to chemotherapy. Cancer Biol Med. 2020;17:842–63.
Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–26.
Alexa-Stratulat T, Pesic M, Gasparovic AC, Trougakos IP, Riganti C. What sustains the multidrug resistance phenotype beyond ABC efflux transporters? Looking beyond the tip of the iceberg. Drug Resist Updat. 2019;46:100643.
Zhu J, Cao K, Zhao M, Ma K, Jiang X, Bai Y, et al. Improvement of ACK1-targeted therapy efficacy in lung adenocarcinoma using chloroquine or bafilomycin A1. Mol Med. 2023;29:6.
Harrington JS, Ryter SW, Plataki M, Price DR, Choi AMK. Mitochondria in health, disease, and aging. Physiol Rev. 2023;103:2349–422.
Vyas S, Zaganjor E, Haigis MC. Mitochondria and cancer. Cell. 2016;166:555–66.
Gustafsson AB, Dorn GW. 2nd. Evolving and expanding the roles of mitophagy as a homeostatic and pathogenic process. Physiol Rev. 2019;99:853–92.
Zhang T, Xue L, Li L, Tang C, Wan Z, Wang R, et al. BNIP3 protein suppresses PINK1 kinase proteolytic cleavage to promote mitophagy. J Biol Chem. 2016;291:21616–29.
Ng MYW, Wai T, Simonsen A. Quality control of the mitochondrion. Dev Cell. 2021;56:881–905.
Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14.
Qiu YH, Zhang TS, Wang XW, Wang MY, Zhao WX, Zhou HM, et al. Mitochondria autophagy: a potential target for cancer therapy. J Drug Target. 2021;29:576–91.
Song C, Pan S, Zhang J, Li N, Geng Q. Mitophagy: a novel perspective for insighting into cancer and cancer treatment. Cell Prolif. 2022;55:e13327.
Lu Y, Li Z, Zhang S, Zhang T, Liu Y, Zhang L. Cellular mitophagy: mechanism, roles in diseases and small molecule pharmacological regulation. Theranostics. 2023;13:736–66.
Tanaka A, Youle RJ. A chemical inhibitor of DRP1 uncouples mitochondrial fission and apoptosis. Mol Cell. 2008;29:409–10.
Nicolli A, Basso E, Petronilli V, Wenger RM, Bernardi P. Interactions of cyclophilin with the mitochondrial inner membrane and regulation of the permeability transition pore, and cyclosporin A-sensitive channel. J Biol Chem. 1996;271:2185–92.
Zhu J, Liu Y, Ao H, Liu M, Zhao M, Ma J. Comprehensive analysis of the immune implication of ACK1 gene in non-small cell lung cancer. Front Oncol. 2020;10:1132.
Tan DS, Haaland B, Gan JM, Tham SC, Sinha I, Tan EH, et al. Bosutinib inhibits migration and invasion via ACK1 in KRAS mutant non-small cell lung cancer. Mol Cancer. 2014;13:13.
Yan C, Li TS. Dual role of mitophagy in cancer drug resistance. Anticancer Res. 2018;38:617–21.
Wang H, Liang Y, Zhang T, Yu X, Song X, Chen Y, et al. C-IGF1R encoded by cIGF1R acts as a molecular switch to restrict mitophagy of drug-tolerant persister tumour cells in non-small cell lung cancer. Cell Death Differ. 2023;30:2365–81.
Glytsou C, Chen X, Zacharioudakis E, Al-Santli W, Zhou H, Nadorp B, et al. Mitophagy promotes resistance to BH3 mimetics in acute myeloid leukemia. Cancer Discov. 2023;13:1656–77.
Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov. 2020;19:39–56.
Xie XQ, Yang Y, Wang Q, Liu HF, Fang XY, Li CL, et al. Targeting ATAD3A-PINK1-mitophagy axis overcomes chemoimmunotherapy resistance by redirecting PD-L1 to mitochondria. Cell Res. 2023;33:215–28.
Liu M, Fan Y, Li D, Han B, Meng Y, Chen F, et al. Ferroptosis inducer erastin sensitizes NSCLC cells to celastrol through activation of the ROS-mitochondrial fission-mitophagy axis. Mol Oncol. 2021;15:2084–105.
Ke S, Zhou T, Yang P, Wang Y, Zhang P, Chen K, et al. Gold nanoparticles enhance TRAIL sensitivity through Drp1-mediated apoptotic and autophagic mitochondrial fission in NSCLC cells. Int J Nanomed. 2017;12:2531–51.
Chen J, Zhou C, Yi J, Sun J, Xie B, Zhang Z, et al. Metformin and arsenic trioxide synergize to trigger Parkin/pink1-dependent mitophagic cell death in human cervical cancer HeLa cells. J Cancer. 2021;12:6310–9.
Galasso C, Nuzzo G, Brunet C, Ianora A, Sardo A, Fontana A, et al. The marine dinoflagellate alexandrium minutum activates a mitophagic pathway in human lung cancer cells. Mar Drugs. 2018;16:502.
Peng HH, Yang HC, Rupa D, Yen CH, Chiu YW, Yang WJ, et al. ACK1 upregulated the proliferation of head and neck squamous cell carcinoma cells by promoting p27 phosphorylation and degradation. J Cell Commun Signal. 2022;16:567–78.
Zhu J, Peng Z, Tian X, Wu T, Sun A, Yang W, et al. Activation of E3 ubiquitin ligase WWP2 by non-receptor tyrosine kinase ACK1. IUBMB Life. 2023;75:595–608.
Wu Y, Hu X, Li Z, Wang M, Li S, Wang X, et al. Transcription factor RFX2 is a key regulator of mouse spermiogenesis. Sci Rep. 2016;6:20435.
Khoyratty TE, Ai Z, Ballesteros I, Eames HL, Mathie S, Martin-Salamanca S, et al. Distinct transcription factor networks control neutrophil-driven inflammation. Nat Immunol. 2021;22:1093–106.
Xu F, Wu Y, Yang Q, Cheng Y, Xu J, Zhang Y, et al. Engineered extracellular vesicles with SHP2 high expression promote mitophagy for Alzheimer’s disease treatment. Adv Mater. 2022;34:e2207107.
Meier JA, Larner AC. Toward a new STATe: the role of STATs in mitochondrial function. Semin Immunol. 2014;26:20–8.
Qu X, Pan P, Cao S, Ma Y, Yang J, Gao H, et al. Immp2l Deficiency Induced Granulosa Cell Senescence Through STAT1/ATF4 Mediated UPR(mt) and STAT1/(ATF4)/HIF1alpha/BNIP3 Mediated Mitophagy: Prevented by Enocyanin. Int J Mol Sci. 2024;25:11122.
Zhu L, Wang Z, Sun X, Yu J, Li T, Zhao H, et al. STAT3/Mitophagy axis coordinates macrophage NLRP3 inflammasome activation and inflammatory bone loss. J Bone Min Res. 2023;38:335–53.
Acknowledgements
This work was supported by the Natural Science Foundation of China [82172786], Heilongjiang Provincial Natural Science Foundation of China [ZD2024H003], Beijing Xisike Clinical Oncology Foundation (Y-2024AZ(EGFR) MS-0187), the Climbing Program of Harbin Medical University Cancer Hospital [PDYS2024-02] and the National Cancer Center Climbing Fund of China [NCC201908B06].
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Data preparation, collection, and analysis were performed by Kui Cao, Shenshui Wei, Tianjiao Ma, Jinhong Zhu, Jianqun Ma, Hongxue Meng, Xinxin Yang, Mengdi Lu, Yuning Wang, Xiangrong He. Jinhong Zhu and Mengdi Lu accessed and verified the underlying data. Reconciliation of underlying data was solved by Jianqun Ma and Jinhong Zhu. The draft of the manuscript was written by Jinhong Zhu, Kui Cao, and Jianqun Ma. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, K., Wei, S., Ma, T. et al. RFX2-BNIP3 axis-driven adaptive mitophagy promotes resistance to ACK1-targeted therapy in non-small cell lung cancer. Oncogene 44, 3461–3475 (2025). https://doi.org/10.1038/s41388-025-03502-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03502-0