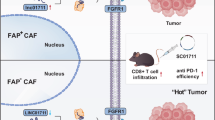
Abstract
The role of cancer-associated fibroblasts (CAFs) in the initiation and invasion phases of human lung adenocarcinoma (LUAD) development is not fully understood. In this study, we utilized single-cell RNA sequencing, spatial transcriptomics, and a combination of in vivo and in vitro models to decode the dynamics of tumor-stroma interactions during human LUAD progression, focusing primarily on adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), and invasive adenocarcinoma (IAC). We identified a matrix CAF (mCAF) subtype characterized by high THBS2 expression, which was closely associated with poor clinical outcomes, tumor recurrence, and the invasive dynamics of LUAD. Spatial transcriptomics and multiplex immunohistochemistry analysis revealed that this CAF subpopulation was closely associated with tumor cells, with clear spatial colocalization. In vivo and in vitro experiments demonstrated that THBS2 secreted by these mCAFs directly binds to SDC4 on tumor cells, enhancing tumor epithelial-mesenchymal transition (EMT) programs. This study highlights THBS2+ mCAFs as key regulators of tumor-stroma interactions and identifies the THBS2-SDC4-EMT axis as a potential therapeutic target in LUAD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All scRNA-seq datasets used in this study are publicly available. The datasets can be accessed via the following links: GSE189357 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189357) and GSE164789 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE164789). The bulk transcriptome data was download from the TCGA (https://portal.gdc.cancer.gov/) and Gene Expression Omnibus database, including GSE14814 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14814), GSE50081 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50081), GSE72094 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72094), GSE31210 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31210), GSE3141 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3141, GSE26939 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26939), GSE30219 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30219), and GSE20913 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20913). Additionally, the 10X Visium Spatial transcriptome data were collected from GSE189487 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE189487).
References
Nicholson AG, Tsao MS, Beasley MB, Borczuk AC, Brambilla E, Cooper WA, et al. The 2021 WHO classification of lung tumors: impact of advances since 2015. J Thorac Oncol. 2022;17:362–87.
Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT. The eighth edition lung cancer stage classification. Chest. 2017;151:193–203.
Yotsukura M, Asamura H, Motoi N, Kashima J, Yoshida Y, Nakagawa K, et al. Long-term prognosis of patients with resected adenocarcinoma in situ and minimally invasive adenocarcinoma of the lung. J Thorac Oncol. 2021;16:1312–20.
Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WE, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:39–51.
Ugalde Figueroa PA, Marques E, Cilento VJ, Giroux DJ, Nishimura KK, Detterbeck FC, et al. Completeness of resection and long-term survival of patients undergoing resection for pathologic T3 NSCLC: an International Association for the Study of Lung Cancer Analysis. J Thorac Oncol. 2024;19:141–52.
Zhu J, Fan Y, Xiong Y, Wang W, Chen J, Xia Y, et al. Delineating the dynamic evolution from preneoplasia to invasive lung adenocarcinoma by integrating single-cell RNA sequencing and spatial transcriptomics. Exp Mol Med. 2022;54:2060–76.
Bremnes RM, Dønnem T, Al-Saad S, Al-Shibli K, Andersen S, Sirera R, et al. The role of tumor stroma in cancer progression and prognosis: emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. J Thorac Oncol. 2011;6:209–17.
Kobayashi H, Gieniec KA, Lannagan TRM, Wang T, Asai N, Mizutani Y, et al. The origin and contribution of cancer-associated fibroblasts in colorectal carcinogenesis. Gastroenterology. 2022;162:890–906.
Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174–86.
Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev. 2014;34:280–300.
Wang Y, Lan W, Xu M, Song J, Mao J, Li C, et al. Cancer-associated fibroblast-derived SDF-1 induces epithelial-mesenchymal transition of lung adenocarcinoma via CXCR4/β-catenin/PPARδ signalling. Cell Death Dis. 2021;12:214.
Liu L, Ba Y, Yang S, Zuo A, Liu S, Zhang Y, et al. FOS-driven inflammatory CAFs promote colorectal cancer liver metastasis via the SFRP1-FGFR2-HIF1 axis. Theranostics. 2025;15:4593–613.
Zhang Y, Zuo A, Ba Y, Liu S, Chen J, Yang S, et al. Cancer-associated fibroblast-derived SEMA3C facilitates colorectal cancer liver metastasis via NRP2-mediated MAPK activation. Proc Natl Acad Sci USA. 2025;122:e2423077122.
Barbazan J, Pérez-González C, Gómez-González M, Dedenon M, Richon S, Latorre E, et al. Cancer-associated fibroblasts actively compress cancer cells and modulate mechanotransduction. Nat Commun. 2023;14:6966.
Lu T, Yang X, Shi Y, Zhao M, Bi G, Liang J, et al. Single-cell transcriptome atlas of lung adenocarcinoma featured with ground glass nodules. Cell Discov. 2020;6:69.
Xing X, Yang F, Huang Q, Guo H, Li J, Qiu M. Decoding the multicellular ecosystem of lung adenocarcinoma manifested as pulmonary subsolid nodules by single-cell RNA sequencing. Sci Adv. 2021;7:eabd9738
Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291.e289.
Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–96.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401.
Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–6.
Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088.
Liu Q, Hsu CY, Shyr Y. Scalable and model-free detection of spatial patterns and colocalization. Genome Res. 2022;32:1736–45.
Liu Z, Deng J, Xu H, Liu L, Zhang Y, Ba Y, et al. Efficient discovery of robust prognostic biomarkers and signatures in solid tumors. Cancer Lett. 2025;613:217502.
Liu Z, Liu L, Weng S, Xu H, Xing Z, Ren Y. et al. BEST: a web application for comprehensive biomarker exploration on large-scale data in solid tumors. J Big Data ;10:165. 2023;10:165
Liu Z, Ba Y, Shan D, Zhou X, Zuo A, Zhang Y, et al. THBS2-producing matrix CAFs promote colorectal cancer progression and link to poor prognosis via the CD47-MAPK axis. Cell Rep. 2025;44:115555.
Cirri P, Chiarugi P. Cancer-associated-fibroblasts and tumour cells: a diabolic liaison driving cancer progression. Cancer Metastasis Rev. 2012;31:195–208.
Yang D, Liu J, Qian H, Zhuang Q. Cancer-associated fibroblasts: from basic science to anticancer therapy. Exp Mol Med. 2023;55:1322–32.
Jin S, Plikus MV, Nie Q. CellChat for systematic analysis of cell-cell communication from single-cell transcriptomics. Nat Protoc. 2025;20:180–219.
Cable DM, Murray E, Zou LS, Goeva A, Macosko EZ, Chen F, et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat Biotechnol. 2022;40:517–26.
Chhabra Y, Weeraratna AT. Fibroblasts in cancer: unity in heterogeneity. Cell. 2023;186:1580–609.
Pelka K, Hofree M, Chen JH, Sarkizova S, Pirl JD, Jorgji V, et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell. 2021;184:4734–4752.e4720.
Li J, Wu C, Hu H, Qin G, Wu X, Bai F, et al. Remodeling of the immune and stromal cell compartment by PD-1 blockade in mismatch repair-deficient colorectal cancer. Cancer Cell. 2023;41:1152–1169.e1157.
Ma C, Yang C, Peng A, Sun T, Ji X, Mi J, et al. Pan-cancer spatially resolved single-cell analysis reveals the crosstalk between cancer-associated fibroblasts and tumor microenvironment. Mol Cancer. 2023;22:170.
Chen C, Guo Q, Liu Y, Hou Q, Liao M, Guo Y, et al. Single-cell and spatial transcriptomics reveal POSTN(+) cancer-associated fibroblasts correlated with immune suppression and tumour progression in non-small cell lung cancer. Clin Transl Med. 2023;13:e1515.
Qin P, Chen H, Wang Y, Huang L, Huang K, Xiao G, et al. Cancer-associated fibroblasts undergoing neoadjuvant chemotherapy suppress rectal cancer revealed by single-cell and spatial transcriptomics. Cell Rep Med. 2023;4:101231.
Kerdidani D, Aerakis E, Verrou KM, Angelidis I, Douka K, Maniou MA. Lung tumor MHCII immunity depends on in situ antigen presentation by fibroblasts. J Exp Med. 2022;219:e20210815
Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9:1102–23.
Kumar L. M EF. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7.
Liao X, Wang W, Yu B, Tan S. Thrombospondin-2 acts as a bridge between tumor extracellular matrix and immune infiltration in pancreatic and stomach adenocarcinomas: an integrative pan-cancer analysis. Cancer Cell Int. 2022;22:213.
Mierke CT. The matrix environmental and cell mechanical properties regulate cell migration and contribute to the invasive phenotype of cancer cells. Rep Prog Phys. 2019;82:064602.
Zhou X, Han J, Zuo A, Ba Y, Liu S, Xu H, et al. THBS2 + cancer-associated fibroblasts promote EMT leading to oxaliplatin resistance via COL8A1-mediated PI3K/AKT activation in colorectal cancer. Mol Cancer. 2024;23:282.
Salmon H, Franciszkiewicz K, Damotte D, Dieu-Nosjean MC, Validire P, Trautmann A, et al. Matrix architecture defines the preferential localization and migration of T cells into the stroma of human lung tumors. J Clin Invest. 2012;122:899–910.
Lavie D, Ben-Shmuel A, Erez N, Scherz-Shouval R. Cancer-associated fibroblasts in the single-cell era. Nat Cancer. 2022;3:793–807.
Cox TR. The matrix in cancer. Nat Rev Cancer. 2021;21:217–38.
Wang X, Eichhorn PJA, Thiery JP. TGF-β, EMT, and resistance to anti-cancer treatment. Semin Cancer Biol. 2023;97:1–11.
Kim HJ, Yang K, Kim K, Lee YJ, Lee S, Ahn SY, et al. Reprogramming of cancer-associated fibroblasts by apoptotic cancer cells inhibits lung metastasis via Notch1-WISP-1 signaling. Cell Mol Immunol. 2022;19:1373–91.
Lawler PR, Lawler J. Molecular basis for the regulation of angiogenesis by thrombospondin-1 and -2. Cold Spring Harb Perspect Med. 2012;2:a006627.
Zhao F, Pu Y, Cui M, Wang H, Cai S. MiR-20a-5p represses the multi-drug resistance of osteosarcoma by targeting the SDC2 gene. Cancer Cell Int. 2017;17:100.
Chute C, Yang X, Meyer K, Yang N, O’Neil K, Kasza I, et al. Syndecan-1 induction in lung microenvironment supports the establishment of breast tumor metastases. Breast Cancer Res. 2018;20:66.
Yao W, Rose JL, Wang W, Seth S, Jiang H, Taguchi A, et al. Syndecan 1 is a critical mediator of macropinocytosis in pancreatic cancer. Nature. 2019;568:410–4.
Karamanos NK, Piperigkou Z, Theocharis AD, Watanabe H, Franchi M, Baud S, et al. Proteoglycan chemical diversity drives multifunctional cell regulation and therapeutics. Chem Rev. 2018;118:9152–232.
Yang H, Liu Y, Zhao MM, Guo Q, Zheng XK, Liu D, et al. Therapeutic potential of targeting membrane-spanning proteoglycan SDC4 in hepatocellular carcinoma. Cell Death Dis. 2021;12:492.
Acknowledgements
This study was supported by the Henan Provincial Science and Technology Research Project (221100310100).
Author information
Authors and Affiliations
Contributions
ZQL conceived, designed, and supervised the research. XWH and DDZ provided project guidance and funding support. YQR and RJM conducted the bioinformatics analysis. ANZ, YHB, STL and HX performed the experiments, with ANZ conducting the statistical analysis of experimental trials. YQR, RJM, and DDZ wrote the manuscript. RJM, PL and QC offered research guidance. YYZ, YY, SYW, JHD, TP, and YKC collected the data. ZQL, RJM and DDZ revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All human tissue samples were obtained with written informed consent from each patient and approved by the Ethics Committee (Approval Nos. HLugA120PG01: 2020-0930; HLugA030PG04-2: 2021-0215; HLugA180Su07: 2022-0401). All animal experiments were approved by the Institutional Animal Care and Use Committee of Zhengzhou University (Ethical Approval No. 2019-KY-383) and conducted in strict accordance with the National Institutes of Health (NIH) guidelines for animal care and use.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ren, Y., Ming, R., Zuo, A. et al. Cancer-associated fibroblasts drive lung adenocarcinoma progression via THBS2-mediated epithelial-mesenchymal transition. Oncogene 44, 4284–4297 (2025). https://doi.org/10.1038/s41388-025-03569-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03569-9
This article is cited by
-
Single-cell multi-omics reveals DUSP9 as a key regulator of cancer stemness and a potential therapeutic target in hepatocellular carcinoma
Journal of Translational Medicine (2026)