Abstract
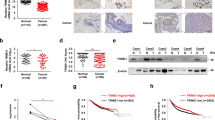
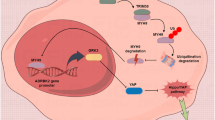
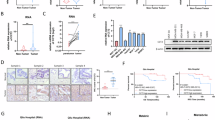
Breast cancer stem cells (BCSCs) are pivotal drivers of breast tumor initiation, metastasis, and therapy resistance. Our previous studies identified the transcription factor SIX2 as a key regulator in maintaining breast cancer stemness. Here, we demonstrate that TRIM21, as an E3 ubiquitin ligase downregulated in breast cancer tissues, binds to SIX2 via its PRY-SPRY domain and catalyzes K48-type ubiquitination at lysine residues K82, K89, and K97. This modification promotes the degradation of SIX2 via the ubiquitin-proteasome pathway, consequently attenuating the stemness and metastatic potential of breast cancer cells. Furthermore, SIX2 transcriptionally activates LGSN expression through direct binding to its promoter region, thereby promoting the stemness and metastatic capabilities of breast cancer cells. Clinically, elevated expression of both SIX2 and LGSN correlates with poor prognosis in breast cancer patients. These results establish that TRIM21-mediated degradation of SIX2 suppresses LGSN expression, ultimately inhibiting the stemness and metastatic abilities of breast cancer cells, underscoring the critical regulatory role of the TRIM21-SIX2-LGSN axis in breast cancer progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
All data are presented within the article and supplementary online data.
References
Liang Y, Zhang H, Song X, Yang Q. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin Cancer Biol. 2020;60:14–27.
Wilkinson L, Gathani T. Understanding breast cancer as a global health concern. Br J Radio. 2022;95:20211033.
Britt KL, Cuzick J, Phillips K-A. Key steps for effective breast cancer prevention. Nat Rev Cancer. 2020;20:417–36.
Zhang L, Chen W, Liu S, Chen C. Targeting Breast Cancer Stem Cells. Int J Biol Sci. 2023;19:552–70.
Taurin S, Alkhalifa H. Breast cancers, mammary stem cells, and cancer stem cells, characteristics, and hypotheses. Neoplasia. 2020;22:663–78.
Crabtree JS, Miele L. Breast Cancer Stem Cells. Biomedicines 2018;6:77.
Nguyen M, Osipo C. Targeting Breast Cancer Stem Cells Using Naturally Occurring Phytoestrogens. Int J Mol Sci. 2022;23:6813.
Zhang R, Tu J, Liu S. Novel molecular regulators of breast cancer stem cell plasticity and heterogeneity. Semin Cancer Biol. 2022;82:11–25.
Okello DO, Iyyanar PPR, Kulyk WM, Smith TM, Lozanoff S, Ji S, et al. Six2 Plays an Intrinsic Role in Regulating Proliferation of Mesenchymal Cells in the Developing Palate. Front Physiol. 2017;8:955.
Pierce J, Murphy AJ, Panzer A, de Caestecker C, Ayers GD, Neblett D, et al. SIX2 Effects on Wilms Tumor Biology. Transl Oncol. 2014;7:800–11.
Zheng L, Guo Q, Xiang C, Liu S, Jiang Y, Gao L, et al. Transcriptional factor six2 promotes the competitive endogenous RNA network between CYP4Z1 and pseudogene CYP4Z2P responsible for maintaining the stemness of breast cancer cells. J Hematol Oncol. 2019;12:23.
Wang C-A, Drasin D, Pham C, Jedlicka P, Zaberezhnyy V, Guney M, et al. Homeoprotein Six2 promotes breast cancer metastasis via transcriptional and epigenetic control of E-cadherin expression. Cancer Res. 2014;74:7357–70.
Velazco-Cruz L, Goedegebuure MM, Maxwell KG, Augsornworawat P, Hogrebe NJ, Millman JR. SIX2 Regulates Human β Cell Differentiation from Stem Cells and Functional Maturation In Vitro. Cell Rep. 2020;31:107687.
Li C, Liu H, Hu Y-C, Lan Y, Jiang R. Generation and characterization of Six2 conditional mice. Genesis. 2020;58:e23365.
Guo Q, Zhou Y, Ni H, Niu M, Xu S, Zheng L, et al. The SIX2/PFN2 feedback loop promotes the stemness of gastric cancer cells. J Transl Med. 2024;22:832.
Liu Z, Li C, Xu J, Lan Y, Liu H, Li X, et al. Crucial and Overlapping Roles of Six1 and Six2 in Craniofacial Development. J Dent Res. 2019;98:572–9.
Oliphant MUJ, Vincent MY, Galbraith MD, Pandey A, Zaberezhnyy V, Rudra P, et al. SIX2 Mediates Late-Stage Metastasis via Direct Regulation of SOX2 and Induction of a Cancer Stem Cell Program. Cancer Res. 2019;79:720–34.
Peng Y, Liu H, Liu J, Long J. Post-translational modifications on mitochondrial metabolic enzymes in cancer. Free Radic Biol Med. 2022;179:11–23.
Yang Y-H, Wen R, Yang N, Zhang T-N, Liu C-F. Roles of protein post-translational modifications in glucose and lipid metabolism: mechanisms and perspectives. Mol Med. 2023;29:93.
Patwardhan A, Cheng N, Trejo J. Post-Translational Modifications of G Protein-Coupled Receptors Control Cellular Signaling Dynamics in Space and Time. Pharmacol Rev. 2021;73:120–51.
Wang Y, Yan D, Liu J, Tang D, Chen X. Protein modification and degradation in ferroptosis. Redox Biol. 2024;75:103259.
Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. 2020;5:11.
Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20:1242–53.
Qin W, Steinek C, Kolobynina K, Forné I, Imhof A, Cardoso MC, et al. Probing protein ubiquitination in live cells. Nucleic Acids Res. 2022;50:e125.
Liu F, Chen J, Li K, Li H, Zhu Y, Zhai Y, et al. Ubiquitination and deubiquitination in cancer: from mechanisms to novel therapeutic approaches. Mol Cancer. 2024;23:148.
Alomari M. TRIM21 - A potential novel therapeutic target in cancer. Pharmacol Res. 2021;165:105443.
Mukadam AS, Miller LVC, Smith AE, Vaysburd M, Sakya SA, Sanford S, et al. Cytosolic antibody receptor TRIM21 is required for effective tau immunotherapy in mouse models. Science. 2023;379:1336–41.
Li J-Y, Zhao Y, Gong S, Wang M-M, Liu X, He Q-M, et al. TRIM21 inhibits irradiation-induced mitochondrial DNA release and impairs antitumour immunity in nasopharyngeal carcinoma tumour models. Nat Commun. 2023;14:865.
Li Y-T, Tan X-Y, Ma L-X, Li H-H, Zhang S-H, Zeng C-M, et al. Targeting LGSN restores sensitivity to chemotherapy in gastric cancer stem cells by triggering pyroptosis. Cell Death Dis. 2023;14:545.
Wyatt K, White HE, Wang L, Bateman OA, Slingsby C, Orlova EV, et al. Lengsin is a survivor of an ancient family of class I glutamine synthetases re-engineered by evolution for a role in the vertebrate lens. Structure. 2006;14:1823–34.
Grassi F, Moretto N, Rivetti C, Cellai S, Betti M, Márquez AJ, et al. Structural and functional properties of lengsin, a pseudo-glutamine synthetase in the transparent human lens. Biochem Biophys Res Commun. 2006;350:424–9.
Nakatsugawa M, Hirohashi Y, Torigoe T, Asanuma H, Takahashi A, Inoda S, et al. Novel spliced form of a lens protein as a novel lung cancer antigen, Lengsin splicing variant 4. Cancer Sci. 2009;100:1485–93.
Nandi A, Chakrabarti R. Assessment of Breast Cancer Stem Cell Activity Using a Spheroid Formation Assay. Methods Mol Biol. 2022;2429:485–500.
Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2024;52:D174–D182.
Győrffy B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation (Camb). 2024;5:100625.
Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27.
Gao W, Li Y, Liu X, Wang S, Mei P, Chen Z, et al. TRIM21 regulates pyroptotic cell death by promoting Gasdermin D oligomerization. Cell Death Differ. 2022;29:439–50.
Cooper J, Giancotti FG. Integrin Signaling in Cancer: Mechanotransduction, Stemness, Epithelial Plasticity, and Therapeutic Resistance. Cancer Cell. 2019;35:347–67.
Kiss L, James LC. The molecular mechanisms that drive intracellular neutralization by the antibody-receptor and RING E3 ligase TRIM21. Semin Cell Dev Biol. 2022;126:99–107.
Pan J-A, Sun Y, Jiang Y-P, Bott AJ, Jaber N, Dou Z, et al. TRIM21 Ubiquitylates SQSTM1/p62 and Suppresses Protein Sequestration to Regulate Redox Homeostasis. Mol Cell. 2016;61:720–33.
Bevacqua RJ, Lam JY, Peiris H, Whitener RL, Kim S, Gu X, et al. SIX2 and SIX3 coordinately regulate functional maturity and fate of human pancreatic β cells. Genes Dev. 2021;35:234–49.
Daino K, Nishimura M, Imaoka T, Takabatake M, Morioka T, Nishimura Y, et al. Epigenetic dysregulation of key developmental genes in radiation-induced rat mammary carcinomas. International J Cancer. 2018;143:343–54.
Balachandran S, Narendran A. The Developmental Origins of Cancer: A Review of the Genes Expressed in Embryonic Cells with Implications for Tumorigenesis. Genes (Basel). 2023;14:604.
Hwang D, Baek S, Chang J, Seol T, Ku B, Ha H, et al. YAP promotes global mRNA translation to fuel oncogenic growth despite starvation. Exp Mol Med. 2024;56:2202–15.
Gwynne WD, Suk Y, Custers S, Mikolajewicz N, Chan JK, Zador Z, et al. Cancer-selective metabolic vulnerabilities in MYC-amplified medulloblastoma. Cancer Cell. 2022;40:1488–1502.e7.
Singh K, Lin J, Zhong Y, Burčul A, Mohan P, Jiang M, et al. c-MYC regulates mRNA translation efficiency and start-site selection in lymphoma. J Exp Med. 2019;216:1509–24.
Wang C, Fan S, Li Z, Fu M, Rao M, Ma Y, et al. Cyclin D1 antagonizes BRCA1 repression of estrogen receptor alpha activity. Cancer Res. 2005;65:6557–67.
Cassandras M, Wang C, Kathiriya J, Tsukui T, Matatia P, Matthay M, et al. Gli1+ mesenchymal stromal cells form a pathological niche to promote airway progenitor metaplasia in the fibrotic lung. Nat Cell Biol. 2020;22:1295–306.
Koli S, Shetty S. Ribosomal dormancy at the nexus of ribosome homeostasis and protein synthesis. Bioessays. 2024;46:e2300247.
Liu Y, Zhang P, Wu Q, Fang H, Wang Y, Xiao Y, et al. Long non-coding RNA NR2F1-AS1 induces breast cancer lung metastatic dormancy by regulating NR2F1 and ΔNp63. Nat Commun. 2021;12:5232.
Popay TM, Wang J, Adams CM, Howard GC, Codreanu SG, Sherrod SD, et al. MYC regulates ribosome biogenesis and mitochondrial gene expression programs through its interaction with host cell factor-1. Elife. 2021;10:e60191.
Li J-W, Huang C-Z, Li J-H, Yuan J-H, Chen Q-H, Zhang W-F, et al. Six2 is negatively correlated with good prognosis and decreases 5-FU sensitivity via suppressing E-cadherin expression in hepatocellular carcinoma cells. Biomed Pharmacother. 2018;104:204–10.
Li J, Cheng X, Huang D, Cui R. Six2 regulates the malignant progression and 5-FU resistance of hepatocellular carcinoma through the PI3K/AKT/mTOR pathway and DNMT1/E-cadherin methylation mechanism. Neoplasma. 2024;71:451–62.
Huang N, Sun X, Li P, Liu X, Zhang X, Chen Q, et al. TRIM family contribute to tumorigenesis, cancer development, and drug resistance. Exp Hematol Oncol. 2022;11:75.
Janciauskiene S, Lechowicz U, Pelc M, Olejnicka B, Chorostowska-Wynimko J. Diagnostic and therapeutic value of human serpin family proteins. Biomed Pharmacother. 2024;175:116618.
Ji B, Harris BRE, Liu Y, Deng Y, Gradilone SA, Cleary MP. et al. Targeting IRES-Mediated p53 Synthesis for Cancer Diagnosis and Therapeutics. Int J Mol Sci. 2017;18:93
Roiuk M, Neff M, Teleman AA. eIF4E-independent translation is largely eIF3d-dependent. Nat Commun. 2024;15:6692
Nalawansha DA, Crews CM. PROTACs: An Emerging Therapeutic Modality in Precision Medicine. Cell Chem Biol 2020;27.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 82204432, 82473955, 82173842), the Science and Technology Research Project of Henan Province (No. 252102311191, No. 242102310257), the Fundamental Research Funds for the Central Universities (2632025TD04), and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Contributions
Lufeng Zheng and Qianqian Guo designed the research. Haitao Chen, Yi Zhou analyzed the data. Haitao Chen, Yi Zhou, Yunnan Zhang, Yannan Fan performed the research. Haitao Chen and Yi Zhou wrote the paper. Lufeng Zheng, Qianqian Guo reviewed this paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All methods were performed in accordance with the relevant guidelines and regulations. All animal care and use procedures were conducted in accordance with the Principles of Animal Care established by China Pharmaceutical University and the Guide for the Care and Use of Laboratory Animals. The study protocol was approved by the Ethics Committee of China Pharmaceutical University (Approval Number: 2025-01-011). All methods were performed in compliance with relevant guidelines and regulations. Informed consent was obtained from all participants involved in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, H., Zhou, Y., Zhang, Y. et al. TRIM21-mediated ubiquitination of SIX2 attenuates breast cancer stemness via LGSN suppression. Oncogene 44, 4173–4189 (2025). https://doi.org/10.1038/s41388-025-03572-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03572-0