Abstract
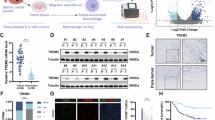
Tumor-associated macrophages (TAMs) play a critical role in promoting tumor progression and represent a promising target for immunotherapeutic intervention. However, the phenotypic characteristics and polarization dynamics of TAMs remain poorly understood, largely due to the complex cellular heterogeneity within the tumor microenvironment. In this study, we comprehensively characterize the heterogeneity of TAMs in gastric adenocarcinoma (GA), with a particular focus on immunosuppressive subsets. Our findings demonstrate that TAMs undergo multidirectional differentiation and exhibit diverse immunoregulatory functions. Among them, FPR3⁺ macrophages are identified as a distinct immunosuppressive population associated with poor patient prognosis. Functional assays using shRNA-mediated knockdown and specific agonists reveal that FPR3 regulates macrophage proliferation and polarization and is essential for TAM formation and maintenance. Mechanistically, FPR3 upregulates FZD7 and CCDC88C, leading to activation of the intracellular Wnt/PCP pathway and downstream JNK signaling, thereby promoting TAM development. Collectively, our study identifies FPR3 as a novel marker of immunosuppressive TAMs, elucidates its mechanistic role in macrophage plasticity, and offers new insights into potential therapeutic strategies for targeting the tumor microenvironment in GA.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data generated in this study are freely available without any restrictions. The raw single cell RNA sequencing data of GA samples have been uploaded to GEO dataset (GSE264203). The bulk RNA-seq raw data of macrophages have been deposited in the Sequence Read Archive (SRA) database (PRJNA1165156). All other data supporting the findings of this research are provided within the article and its supplementary materials. The external raw data that support the findings were obtained from Gene Expression Omnibus (GEO) at GSE183904, GSE15459 and TCGA STAD project (https://gdc.cancer.gov).
References
Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov. 2018;17:887–904.
Li J, Sun J, Zeng Z, Liu Z, Ma M, Zheng Z, et al. Tumour-associated macrophages in gastric cancer: from function and mechanism to application. Clin Transl Med. 2023;13:e1386.
Bleriot C, Dunsmore G, Alonso-Curbelo D, Ginhoux F. A temporal perspective for tumor-associated macrophage identities and functions. Cancer Cell. 2024;42:747–58.
Xiang X, Wang J, Lu D, Xu X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Signal Transduct Target Ther. 2021;6:75.
Sathe A, Grimes SM, Lau BT, Chen J, Suarez C, Huang RJ, et al. Single-cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin Cancer Res. 2020;26:2640–53.
Dong Y, Hu K, Zhang J, Zhu M, Liu M, Yuan Y, et al. ScRNA-seq of gastric cancer tissues reveals differences in the immune microenvironment of primary tumors and metastases. Oncogene. 2024;43:1549–64.
Kang B, Camps J, Fan B, Jiang H, Ibrahim MM, Hu X, et al. Parallel single-cell and bulk transcriptome analyses reveal key features of the gastric tumor microenvironment. Genome Biol. 2022;23:265.
Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184:792–809.e723.
Chen K, Bao Z, Gong W, Tang P, Yoshimura T, Wang JM. Regulation of inflammation by members of the formyl-peptide receptor family. J Autoimmun. 2017;85:64–77.
Panaro MA, Acquafredda A, Sisto M, Lisi S, Maffione AB, Mitolo V. Biological role of the N-formyl peptide receptors. Immunopharmacol Immunotoxicol. 2006;28:103–27.
Migeotte I, Riboldi E, Franssen JD, Gregoire F, Loison C, Wittamer V, et al. Identification and characterization of an endogenous chemotactic ligand specific for FPRL2. J Exp Med. 2005;201:83–93.
Devosse T, Guillabert A, D’Haene N, Berton A, De Nadai P, Noel S, et al. Formyl peptide receptor-like 2 is expressed and functional in plasmacytoid dendritic cells, tissue-specific macrophage subpopulations, and eosinophils. J Immunol. 2009;182:4974–84.
Birkl D, O’Leary MN, Quiros M, Azcutia V, Schaller M, Reed M, et al. Formyl peptide receptor 2 regulates monocyte recruitment to promote intestinal mucosal wound repair. FASEB J. 2019;33:13632–43.
Klaver D, Posch B, Geisler A, Hermann M, Reider N, Heufler C. Peptides from allergenic lipocalins bind to formyl peptide receptor 3 in human dendritic cells to mediate TH2 immunity. J Allergy Clin Immunol. 2020;145:654–65.
Vital SA, Becker F, Holloway PM, Russell J, Perretti M, Granger DN, et al. Formyl-peptide receptor 2/3/lipoxin A4 receptor regulates neutrophil-platelet aggregation and attenuates cerebral inflammation: impact for therapy in cardiovascular disease. Circulation. 2016;133:2169–79.
Qi J, Liu Y, Hu J, Lu L, Dou Z, Dai H, et al. Identification of FPR3 as a Unique Biomarker for Targeted Therapy in the Immune Microenvironment of Breast Cancer. Front Pharm. 2020;11:593247.
Wu L, Wu W, Zhang J, Zhao Z, Li L, Zhu M, et al. Natural coevolution of tumor and immunoenvironment in glioblastoma. Cancer Discov. 2022;12:2820–37.
Zhou Q, Yan X, Guo Y, Jiang X, Cao T, Ke Y. Machine learning algorithms for predicting glioma patient prognosis based on CD163+FPR3+ macrophage signature. npj Precision Oncol. 2024;8:201.
Tan Z, Pan K, Sun M, Pan X, Yang Z, Chang Z, et al. CCKBR+ cancer cells contribute to the intratumor heterogeneity of gastric cancer and confer sensitivity to FOXO inhibition. Cell Death Differ. 2024;31:1302–17.
Feng M, Jiang W, Kim BYS, Zhang CC, Fu YX, Weissman IL. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat Rev Cancer. 2019;19:568–86.
Mantovani A, Allavena P, Marchesi F, Garlanda C. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discov. 2022;21:799–820.
Lin S, Dai Y, Han C, Han T, Zhao L, Wu R, et al. Single-cell transcriptomics reveal distinct immune-infiltrating phenotypes and macrophage-tumor interaction axes among different lineages of pituitary neuroendocrine tumors. Genome Med. 2024;16:60.
Tran N, Mills EL. Redox regulation of macrophages. Redox Biol. 2024;72:103123.
Kersten K, Hu KH, Combes AJ, Samad B, Harwin T, Ray A, et al. Spatiotemporal co-dependency between macrophages and exhausted CD8(+) T cells in cancer. Cancer Cell. 2022;40:624–38.e629.
Arensman MD, Kovochich AN, Kulikauskas RM, Lay AR, Yang PT, Li X, et al. WNT7B mediates autocrine Wnt/beta-catenin signaling and anchorage-independent growth in pancreatic adenocarcinoma. Oncogene. 2014;33:899–908.
Ferrari ME, Bernis ME, McLeod F, Podpolny M, Coullery RP, Casadei IM, et al. Wnt7b signalling through Frizzled-7 receptor promotes dendrite development by coactivating CaMKII and JNK. J Cell Sci. 2018;131:jcs216101.
Aznar N, Sun N, Dunkel Y, Ear J, Buschman MD, Ghosh P. A Daple-Akt feed-forward loop enhances noncanonical Wnt signals by compartmentalizing beta-catenin. Mol Biol Cell. 2017;28:3709–23.
Aznar N, Ear J, Dunkel Y, Sun N, Satterfield K, He F, et al. Convergence of Wnt, growth factor, and heterotrimeric G protein signals on the guanine nucleotide exchange factor Daple. Sci Signal. 2018;11:eaao4220.
Esaki N, Enomoto A, Takagishi M, Mizutani Y, Iida T, Ushida K, et al. The Daple-CK1epsilon complex regulates Dvl2 phosphorylation and canonical Wnt signaling. Biochem Biophys Res Commun. 2020;532:406–13.
Ishida-Takagishi M, Enomoto A, Asai N, Ushida K, Watanabe T, Hashimoto T, et al. The Dishevelled-associating protein Daple controls the non-canonical Wnt/Rac pathway and cell motility. Nat Commun. 2012;3:859.
Xie N, Landin Malt A, Adylkhan A, Rodeman N, Moraes Borges R, Hwang D, et al. Wnt7b acts in concert with Wnt5a to regulate tissue elongation and planar cell polarity via noncanonical Wnt signaling. Proc Natl Acad Sci USA. 2024;121:e2405217121.
Deng Z, Loyher PL, Lazarov T, Li L, Shen Z, Bhinder B, et al. The nuclear factor ID3 endows macrophages with a potent anti-tumour activity. Nature. 2024;626:864–73.
Cai Z, Li W, Hager S, Wilson JL, Afjehi-Sadat L, Heiss EH, et al. Targeting PHGDH reverses the immunosuppressive phenotype of tumor-associated macrophages through alpha-ketoglutarate and mTORC1 signaling. Cell Mol Immunol. 2024;21:448–65.
He HQ, Ye RD. The formyl peptide receptors: diversity of ligands and mechanism for recognition. Molecules. 2017;22:455.
Cuomo P, Papaianni M, Capparelli R, Medaglia C. The role of formyl peptide receptors in permanent and low-grade inflammation: helicobacter pylori infection as a model. Int J Mol Sci. 2021;22:3706.
Prevete N, Liotti F, Marone G, Melillo RM, de Paulis A. Formyl peptide receptors at the interface of inflammation, angiogenesis and tumor growth. Pharm Res. 2015;102:184–91.
Wang H, Wang X, Zhang X, Xu W. The promising role of tumor-associated macrophages in the treatment of cancer. Drug Resist Updat. 2024;73:101041.
Dalla E, Papanicolaou M, Park MD, Barth N, Hou R, Segura-Villalobos D, et al. Lung-resident alveolar macrophages regulate the timing of breast cancer metastasis. Cell. 2024;187:6631–48.e6620.
Rabiet MJ, Macari L, Dahlgren C, Boulay F. N-formyl peptide receptor 3 (FPR3) departs from the homologous FPR2/ALX receptor with regard to the major processes governing chemoattractant receptor regulation, expression at the cell surface, and phosphorylation. J Biol Chem. 2011;286:26718–31.
Acknowledgements
We express gratitude to all members of the Pathophysiology Department at Anhui Medical University for their valuable comments and advice. Additionally, we acknowledge the Center for Scientific Research at Anhui Medical University for their invaluable support in conducting our experiments. We thank OE Biotech Co., Ltd (Shanghai, China) for providing single-cell RNA-seq and assistance with bioinformatics analysis.
Funding
This work was supported by the National Natural Science Foundation of China (reference numbers 82173004, 81870085,81670097), Chinese Scholarship Council Research Collaboration Project, Grants for Scientific Research Enhancement of Anhui Medical University (2019xkjT004 and XJ2020019), Grants for Collaborative Innovation Project of Colleges and Universities in Anhui Province (GXXT-2021-063), Anhui Medical University Basic and Clinical Cooperation Project (2022xkjT029), Natural Science Foundation of Anhui Province Education Department (2022AH051183), Postgraduate Innovation Research and Practice Program of Anhui Medical University (YJS20230101, YJS20230144).
Author information
Authors and Affiliations
Contributions
All coauthors have contributed to this study. MS, ZT, and KL conducted the experiments, analyzed data, and wrote this paper. JZ and YL were responsible for biopsies collection and data collection. ZC and ZY interpretated scRNA-seq data. ZC, ZY, XY, GT, and CL were responsible for biological experiments and manuscript revision. HZ, CP, and CK were responsible for experimental design, data interpretation, and manuscript revision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
All studies and methods were conducted in accordance with the regulations and guidelines approved by the Ethics Committee of Anhui Medical University. The study received approval from the ethics committee of Anhui Medical University (LLSC20210872). Written informed consent was obtained from all study participants. All the animal experiments of this study were approved by the Animal Care and Use Committees at Anhui Medical University (LLSC20210972).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, M., Tan, Z., Lin, K. et al. FPR3 sustains the immunosuppression of tumor-associated macrophages and accelerates the progression of gastric adenocarcinoma. Oncogene 44, 4309–4323 (2025). https://doi.org/10.1038/s41388-025-03578-8
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03578-8