Abstract
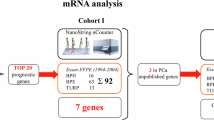
Aging significantly influences the pathogenesis of prostate cancer (PCa). Emerging evidence suggests that aging-related methylation changes play a critical role in PCa. However, the impact of aging-related DNA methylation in PCa remains largely unexplored. To identify hypermethylated sites associated with aging in PCa, we performed an epigenome-wide analysis using Illumina Human Methylation BeadChip arrays. The candidate methylation markers were further refined through least absolute shrinkage and selection operator (LASSO) regression and Random Forest model. Besides, we investigate the functional role of ZNF573 in PCa. Our analysis identified four aging-related CpG sites in the promoter region of ZNF573 that exhibited significant hypermethylation in PCa. These four DNA methylation markers effectively distinguished PCa from benign prostatic hyperplasia (BPH) with high AUC (0.847), which was superior to PSA. Furthermore, the expression of ZNF573 was notably down-regulated in PCa, and its overexpression significantly inhibited PCa cells proliferation and invasion both in vivo and in vitro. ZNF573 acting as a transcription factor promoted the expression of the E3 ubiquitin ligase RNF19B, which regulated the ubiquitination of PIK3CA. These findings suggest that aging-related ZNF573 methylation could serve as a potential diagnostic biomarker for PCa, influencing the development and progression of PCa through the regulation of PIK3CA ubiquitination via RNF19B.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data used and/or analyzed in this study are available on request from the corresponding author.
References
Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63.
Bergengren O, Pekala KR, Matsoukas K, Fainberg J, Mungovan SF, Bratt O, et al. Update on prostate cancer epidemiology and risk factors—a systematic review. Eur Urol. 2023;84:191–206. 2022.
Behr LC, Simm A, Kluttig A, Grosskopf A. Grosskopf, 60 years of healthy aging: on definitions, biomarkers, scores and challenges. Ageing Res Rev. 2023;88:101934.
Liu X, Yu C, Bi Y, Zhang ZJ. Trends and age-period-cohort effect on incidence and mortality of prostate cancer from 1990 to 2017 in China. Public Health. 2019;172:70–80.
Welch HG, Albertsen PC. Reconsidering prostate cancer mortality - the future of PSA screening. N Engl J Med. 2020;382:1557–63.
Loeb S, van den Heuvel S, Zhu X, Bangma CH, Schroder FH, Roobol MJ. Infectious complications and hospital admissions after prostate biopsy in a European randomized trial. Eur Urol. 2012;61:1110–4.
Lopez-Otin C, Pietrocola F, Roiz-Valle D, Galluzzi L, Kroemer G. Meta-hallmarks of aging and cancer. Cell Metab. 2023;35:12–35.
Mc Auley MT. DNA methylation in genes associated with the evolution of ageing and disease: a critical review. Ageing Res Rev. 2021;72:101488.
Field AE, Robertson NA, Wang T, Havas A, Ideker T, Adams PD. DNA methylation clocks in aging: categories, causes, and consequences. Mol Cell. 2018;71:882–95.
Li J, Zhu X, Yu K, Jiang H, Zhang Y, Wang B, et al. Exposure to polycyclic aromatic hydrocarbons and accelerated DNA methylation aging. Environ Health Perspect. 2018;126:067005.
Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–6.
Yang PL, Lai TS, Chou YH, Lai LC, Lin SL, Chen YM. DNA methylation in peripheral blood is associated with renal aging and renal function decline: a national community study. Clin Epigenet. 2024;16:80.
Allen PC, Roberts K, Rubio JE, Tiwari HK, Absher DM, Cooper SJ, et al. Genome-wide DNA methylation analysis implicates enrichment of interferon pathway in African American patients with Systemic Lupus Erythematosus and European Americans with lupus nephritis. J Autoimmun. 2023;139:103089.
Ma G, Liu H, Hua Q, Wang M, Du M, Lin Y, et al. KCNMA1 cooperating with PTK2 is a novel tumor suppressor in gastric cancer and is associated with disease outcome. Mol Cancer. 2017;16:46.
Che L, Huang J, Lin JX, Xu CY, Wu XM, Du ZB, et al. Aflatoxin B1 exposure triggers hepatic lipotoxicity via p53 and perilipin 2 interaction-mediated mitochondria-lipid droplet contacts: an in vitro and in vivo assessment. J Hazard Mater. 2023;445:130584.
Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–67.
Aref-Eshghi E, Schenkel LC, Ainsworth P, Lin H, Rodenhiser DI, Cutz JC, et al. Genomic DNA methylation-derived algorithm enables accurate detection of malignant prostate tissues. Front Oncol. 2018;8:100.
Kirby MK, Ramaker RC, Roberts BS, Lasseigne BN, Gunther DS, Burwell TC, et al. Genome-wide DNA methylation measurements in prostate tissues uncovers novel prostate cancer diagnostic biomarkers and transcription factor binding patterns. BMC Cancer. 2017;17:273.
Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560.
Oughtred R, Rust J, Chang C, Breitkreutz BJ, Stark C, Willems A, et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30:187–200.
Ying Y, Wang M, Chen Y, Li M, Ma C, Zhang J, et al. Zinc finger protein 280C contributes to colorectal tumorigenesis by maintaining epigenetic repression at H3K27me3-marked loci. Proc Natl Acad Sci USA. 2022;119:e2120633119.
Martins F, Rosspopoff O, Carlevaro-Fita J, Forey R, Offner S, Planet E, et al. A cluster of evolutionarily recent krab zinc finger proteins protects cancer cells from replicative stress-induced inflammation. Cancer Res. 2024;84:808–26.
Arafeh R, Samuels Y. PIK3CA in cancer: the past 30 years. Semin Cancer Biol. 2019;59:36–49.
de Magalhaes JP. How ageing processes influence cancer. Nat Rev Cancer. 2013;13:357–65.
Perez RF, Tejedor JR, Fernandez AF, Fraga MF. Aging and cancer epigenetics: where do the paths fork? Aging Cell. 2022;21:e13709.
Chatsirisupachai K, Palmer D, Ferreira S, de Magalhaes JP. A human tissue-specific transcriptomic analysis reveals a complex relationship between aging, cancer, and cellular senescence. Aging Cell. 2019;18:e13041.
Chatsirisupachai K, Lesluyes T, Paraoan L, Van Loo P, de Magalhaes JP. An integrative analysis of the age-associated multi-omic landscape across cancers. Nat Commun. 2021;12:2345.
Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8:e1002629.
Yousefi PD, Suderman M, Langdon R, Whitehurst O, Smith GDavey, Relton CL. DNA methylation-based predictors of health: applications and statistical considerations. Nat Rev Genet. 2022;23:369–83.
Wang K, Liu H, Hu Q, Wang L, Liu J, Zheng Z, et al. Epigenetic regulation of aging: implications for interventions of aging and diseases. Signal Transduct Target Ther. 2022;7:374.
Bell CG, Lowe R, Adams PD, Baccarelli AA, Beck S, Bell JT, et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol. 2019;20:249.
Izadi M, Sadri N, Abdi A, Serajian S, Jalalei D, Tahmasebi S. Epigenetic biomarkers in aging and longevity: current and future application. Life Sci. 2024;351:122842.
Zabransky DJ, Jaffee EM, Weeraratna AT. Shared genetic and epigenetic changes link aging and cancer. Trends Cell Biol. 2022;32:338–50.
Xie W, Kagiampakis I, Pan L, Zhang YW, Murphy L, Tao Y, et al. DNA methylation patterns separate senescence from transformation potential and indicate cancer risk. Cancer Cell. 2018;33:309–21.e305.
Kwabi-Addo B, Chung W, Shen L, Ittmann M, Wheeler T, Jelinek J, et al. Age-related DNA methylation changes in normal human prostate tissues. Clin Cancer Res. 2007;13:3796–802.
Kumaraswamy A, Welker Leng KR, Westbrook TC, Yates JA, Zhao SG, Evans CP, et al. Recent advances in epigenetic biomarkers and epigenetic targeting in prostate cancer. Eur Urol. 2021;80:71–81.
Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, Consortium C. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31:745–59.
Constancio V, Nunes SP, Moreira-Barbosa C, Freitas R, Oliveira J, Pousa I, et al. Early detection of the major male cancer types in blood-based liquid biopsies using a DNA methylation panel. Clin Epigenetics. 2019;11:175.
Lomniczi A, Wright H, Castellano JM, Matagne V, Toro CA, Ramaswamy S, et al. Epigenetic regulation of puberty via Zinc finger protein-mediated transcriptional repression. Nat Commun. 2015;6:10195.
Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci. 2016;23:53.
Li X, Han M, Zhang H, Liu F, Pan Y, Zhu J, et al. Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma. Biomark Res. 2022;10:2.
Hoover RG, Gullickson G, Kornbluth J. Natural killer lytic-associated molecule plays a role in controlling tumor dissemination and metastasis. Front Immunol. 2012;3:393.
Hoover RG, Gullickson G, Kornbluth J. Impaired NK cytolytic activity and enhanced tumor growth in NK lytic-associated molecule-deficient mice. J Immunol. 2009;183:6913–21.
Li J, Wang H, Lu Q, Han J, Xu H, Sun P. Lysosome-related genes and RNF19 b as prognostic markers for survival and immunotherapy efficacy in hepatocellular carcinoma. Clin Transl Gastroenterol. 2024;15:e1.
Wang Y, Wei M, Su M, Du Z, Dong J, Zhang Y, et al. DIRAS3 enhances RNF19B-mediated RAC1 ubiquitination and degradation in non-small-cell lung cancer cells. iScience. 2023;26:107157.
Tortorella E, Giantulli S, Sciarra A, Silvestri I. In prostate cancer: a tale of two interconnected pathways. Int J Mol Sci. 2023;24:2046.
Zhou Y, Xu J, Luo H, Meng X, Chen M, Zhu D. Wnt signaling pathway in cancer immunotherapy. Cancer Lett. 2022;525:84–96.
Xue C, Chu Q, Shi Q, Zeng Y, Lu J, Li L. Wnt signaling pathways in biology and disease: mechanisms and therapeutic advances. Signal Transduct Target Ther. 2025;10:106.
Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–47.
Soutto M, Peng D, Katsha A, Chen Z, Piazuelo MB, Washington MK, et al. Activation of beta-catenin signalling by TFF1 loss promotes cell proliferation and gastric tumorigenesis. Gut. 2015;64:1028–39.
Li H, Liu S, Jin R, Xu H, Li Y, Chen Y, et al. Pyrvinium pamoate regulates MGMT expression through suppressing the Wnt/beta-catenin signaling pathway to enhance the glioblastoma sensitivity to temozolomide. Cell Death Discov. 2021;7:288.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (JUSRP11951).
Author information
Authors and Affiliations
Contributions
Study conception and design: GM, YG. Acquisition and analysis of data: YG, PH, YS. Drafting and revision of the manuscript: YG, GM, LC, QY. Administrative, technical or material support: YC, QS, LC, QY. Study supervision: GM. Final approval of the manuscript: all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All subjects provided written informed consent prior to enrollment. The study protocol was approved by the Institutional Review Board of the Third Affiliated Hospital of Soochow University (CL022H71-01). All animal experiments were performed in accordance with protocols approved by the Animal Care Committee of Jiangnan University and in compliance with the guidelines set by the Institutional Animal Care and Use Committee (JN. No20230215b0320915[025]).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ge, Y., Han, P., Sun, Y. et al. Aging-associated ZNF573 methylation regulates RNF19B-PIK3CA ubiquitination to promote prostate cancer. Oncogene (2025). https://doi.org/10.1038/s41388-025-03579-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41388-025-03579-7