Abstract
Background
Genome-wide association studies (GWAS) in healthy populations have identified variants associated with erythrocyte traits, but genetic causes of hemoglobin variation in children with CKD are incompletely understood.
Methods
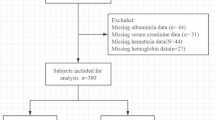
The Pediatric Investigation of Genetic Factors Linked with Renal Progression (PediGFR) Consortium comprises three pediatric CKD cohorts: Chronic Kidney Disease in Children (CKiD), Effect of Strict Blood Pressure Control and ACE Inhibition on the Progression of CRF in Pediatric Patients (ESCAPE), and Cardiovascular Comorbidity in Children with CKD (4C). We performed cross-sectional and longitudinal association studies of single-nucleotide polymorphisms (SNPs) in 1125 patients.
Results
Children of European (n = 725) or Turkish (n = 400) ancestry (EA or TA) were included. In cross-sectional analysis, two SNPs (rs10758658 and rs12718597) previously associated with RBC traits were significantly associated with hemoglobin levels in children of EA and TA. In longitudinal analysis, SNP rs2540917 was nominally associated with hemoglobin in EA and TA children.
Conclusions
SNPs associated with erythrocyte traits in healthy populations were marginally significant for an association with hemoglobin. Further analyses/replication studies are needed in larger CKD cohorts to investigate SNPs of unknown significance associated with hemoglobin. Functional studies will be required to confirm that the observed associations between SNPs and clinical phenotype are causal.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Atkinson, M. A. & Furth, S. L. Anemia in children with chronic kidney disease. Nat. Rev. Nephrol. 7, 635–641 (2011).
Fadrowski, J. J. et al. Hemoglobin decline in children with chronic kidney disease: baseline results from the chronic kidney disease in children prospective cohort study. Clin. J. Am. Soc. Nephrol. 3, 457–462 (2008).
Borzych-Duzalka, D. et al. Management of anemia in children receiving chronic peritoneal dialysis. J. Am. Soc. Nephrol. 24, 665–676 (2013).
Atkinson, M. A. et al. Hemoglobin differences by race in children with CKD. Am. J. Kidney Dis. 55, 1009–1017 (2010).
Robins, E. B. & Blum, S. Hematologic reference values for African American children and adolescents. Am. J. Hematol. 82, 611–614 (2007).
Ganesh, S. K. et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat. Genet. 41, 1191–1198 (2009).
Li, J. et al. GWAS of blood cell traits identifies novel associated loci and epistatic interactions in Caucasian and African-American Children. Hum. Mol. Genet. 22, 1457–1464 (2013).
Ding, K. et al. Genetic variants that confer resistance to malaria are associated with red blood cell traits in African-Americans: an electronic medical record-based genome-wide association study. G3 (Bethesda) 3, 1061–1068 (2013).
Atkinson, M. A., Martz, K., Warady, B. A. & Neu, A. M. Risk for anemia in pediatric chronic kidney disease patients: a report of NAPRTCS. Pediatr. Nephrol. 25, 1699–1706 (2010).
Pfeffer, M. A. et al. A trial of darbepoetin alfa in type 2 diabetes and chronic kidney disease. N. Engl. J. Med. 361, 2019–2032 (2009).
Singh, A. K. et al. Correction of anemia with epoetin alfa in chronic kidney disease. N. Engl. J. Med. 355, 2085–2098 (2006).
Lestz, R. M., Fivush, B. A. & Atkinson, M. A. Association of higher erythropoiesis stimulating agent dose and mortality in children on dialysis. Pediatr. Nephrol. 29, 2021–2028 (2014).
Wuttke, M. et al. Genetic loci associated with renal function measures and chronic kidney disease in children: the Pediatric Investigation for Genetic Factors Linked with Renal Progression Consortium. Nephrol. Dial. Transplant. 31, 262–269 (2016).
Gupta, J. et al. Genome-wide association studies in pediatric chronic kidney disease. Pediatr. Nephrol. 31, 1241–1252 (2016).
Furth, S. L. et al. Design and methods of the Chronic Kidney Disease in Children (CKiD) prospective cohort study. Clin. J. Am. Soc. Nephrol. 1, 1006–1015 (2006).
ESCAPE Trial Group, Wühl, E. et al. Strict blood-pressure control and progression of renal failure in children. N. Engl. J. Med. 361, 1639–1650 (2009).
Querfeld, U. et al. The Cardiovascular Comorbidity in Children with Chronic Kidney Disease (4C) study: objectives, design, and methodology. Clin. J. Am. Soc. Nephrol. 5, 1642–1648 (2010).
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010).
Delaneau, O., Zagury, J. F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6 (2013).
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
1000 Genomes Project Consortium, Abecasis et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Schwartz, G. J. et al. New equations to estimate GFR in children with CKD. J. Am. Soc. Nephrol. 20, 629–637 (2009).
Arnold, M., Raffler, J., Pfeufer, A., Suhre, K. & Kastenmüller, G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics 31, 1334–1336 (2015).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Ulirsch, J. C. et al. Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell 165, 1530–1545 (2016).
Sarma, P. R. in Clinical Methods: The History, Physical, and Laboratory Examinations 3rd edn (eds Walker, H. K., Hall, W. D. & Hurst, J. W.) Ch. 152 (Butterworths, Boston, 1990).
Fanis, P., Kousiappa, I., Phylactides, M. & Kleanthous, M. Genotyping of BCL11A and HBS1L-MYB SNPs associated with fetal haemoglobin levels: a SNaPshot minisequencing approach. BMC Genom. 15, 108 (2014).
Basak, A. & Sankaran, V. G. Regulation of the fetal hemoglobin silencing factor BCL11A. Ann. NY Acad. Sci. 1368, 25–30 (2016).
Marian, A. J. The enigma of genetics etiology of atherosclerosis in the post-GWAS era. Curr. Atheroscler. Rep. 14, 295–299 (2012).
Singleton, A. B. et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science 302, 841 (2003).
Tan, E. K. et al. Alpha synuclein promoter and risk of Parkinson’s disease: microsatellite and allelic size variability. Neurosci. Lett. 36, 70–72 (2003).
Schneider, J. A. et al. DNA variability of human genes. Mech. Ageing Dev. 124, 17–25 (2003).
Nielsen, R., Hubisz, M. J. & Clark, A. G. Reconstituting the frequency spectrum of ascertained single-nucleotide polymorphism data. Genetics 168, 2373–2382 (2004).
Lin, P.-I., Vance, J. M., Pericak-Vance, M. A. & Martin, E. R. No gene is an island: the flip-flop phenomenon. Am. J. Hum. Genet. 80, 531–538 (2007).
Amos, W., Driscoll, E. & Hoffman, J. I. Candidate genes versus genome-wide associations: which are better for detecting genetic susceptibility to infectious disease? Proc. Biol. Sci. 278, 1183–1188 (2011).
Acknowledgements
The CKiD Study is supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), with additional funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and the National Heart, Lung, and Blood Institute (U01-DK-66143, U01-DK-66174, U01DK-082194, and U01-DK-66116). The CKID website is located at http://www.statepi.jhsph.edu/ckid. The ESCAPE trial and the 4C Study were conducted by the ESCAPE Clinical Research Network. The ESCAPE Trial was supported by grants from the European Commission Fifth Framework Programme (QLRT-2001-00908), Boehringer Ingelheim Foundation, Kuratorium für Dialyse und Nierentransplantation e.V., and the Baxter Extramural Grant Program. Support for the 4C Study was received from the ERA-EDTA Research Programme, the KfH Foundation for Preventive Medicine and the German Federal Ministry of Education and Research (reference number: 01EO0802). The genotyping data utilized for the PediGFR Consortium studies (CKiD, 4C, and ESCAPE) were supported by NIDDK Grant No. RO1-DK-082394. Additional support was provided by the EU 7th Framework Programme (EURenOmics, Grant 2012-305608). M.A.A. was supported by the National Institutes of Health (NIH)/NIDDK (K23-DK-084116). The work of M.W. and A.K was supported by the CRC 1140 of the German Research Foundation. M.A.A. and S.L.F. were supported by an unrestricted grant from Amgen, Inc.
Author information
Authors and Affiliations
Contributions
M.A.A participated in the analysis and interpretation of data, drafted and revised the article providing intellectual content of critical importance to the work, and approved the final draft. R.X. participated in the analysis and interpretation of the data, revised the article providing intellectual content of critical importance to the work, and approved the final draft. A.K. participated in the conception and design of the analysis and the interpretation of data, revised the article and provided intellectual content of critical importance to the work, and approved the final draft. E.W. participated in the conception and design of the analysis and in the analysis and interpretation of data, revised the article and provided intellectual content of critical importance to the work, and approved the final draft. C.S.W. participated in the conception and design of the analysis and in the analysis and interpretation of data, revised the article and provided intellectual content of critical importance to the work, and approved the final draft. M.W. participated in the conception and design of the analysis and in the analysis and interpretation of data, revised the article and provided intellectual content of critical importance to the work, and approved the final draft. A.K.B. participated in the design of the analysis, revised the article for critical intellectual content, and approved the final draft. S.Ç. participated in the design of the analysis, revised the article for critical intellectual content, and approved the final draft. B.A.W. participated in the conception and design of the analysis and in the interpretation of data, revised the article and provided intellectual content of critical importance to the work, and approved the final draft. F.S. participated in the conception and design of the analysis and in the interpretation of data, revised the article and provided intellectual content of critical importance to the work, and approved the final draft. S.L.F. participated in the conception and design of the analysis and in the interpretation of data, revised the article and provided intellectual content of critical importance to the work, and approved the final draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Atkinson, M.A., Xiao, R., Köttgen, A. et al. Genetic associations of hemoglobin in children with chronic kidney disease in the PediGFR Consortium. Pediatr Res 85, 324–328 (2019). https://doi.org/10.1038/s41390-018-0148-z
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41390-018-0148-z
This article is cited by
-
Clinical profile of children attending the pediatric nephrology outpatient clinic: a cross section study
Egyptian Pediatric Association Gazette (2025)