Abstract
Background
Hirschsprung’s disease (HSCR) is the most common congenital cause of intestinal obstruction in children. Sotos syndrome (SoS) is an overgrowth disorder with constipation and sometimes accompanied by HSCR. NSD1 gene mutation is the main cause of SoS. We aimed to investigate association of NSD1 common single nucleotide polymorphisms (SNPs) with HSCR susceptibility in Chinese Han population.
Method
We genotyped 15 SNPs encompassing NSD1 gene region in 420 HSCR patients and 1665 controls on Fludigm EP1 platform. Association analysis was performed between cases and controls.
Result
Rs244709 was the most associated SNP with HSCR susceptibility of the sample set (PAllelic = 9.69 × 10−5, OR = 1.37, 95% CI: 1.17–1.61). Gender stratification analysis revealed that NSD1 SNPs were associated with HSCR in males, but not in females. The nonsynonymous coding SNP rs28932178 in NSD1 exon 5 represented the most significant signal in males (PAllelic = 6.43 × 10−5, OR = 1.42, 95% CI: 1.20–1.69). The associated SNPs were expression quantitative trait loci (eQTLs) of nearby genes in multiple tissues. NSD1 expression levels were higher in aganglionic colon tissues than ganglionic tissues (P = 3.00 × 10−6).
Conclusion
NSD1 variation conferred risk to HSCR in males, indicating SoS and HSCR may share common genetic factors.
Impact
-
This is the first study to reveal that NSD1 variation conferred risk to Hirschsprung’s disease susceptibility in males of Chinese Han population, indicating Sotos syndrome and Hirschsprung’s disease may share some common genetic background.
-
This study indicates more attention should be paid to the symptom of constipation in patients with Sotos syndrome.
-
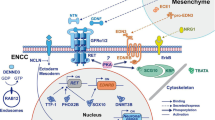
Our results raise questions about the role of NSD1 in the development of enteric nervous system and the pathogenesis of Hirschsprung’s disease.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Haricharan, R. N. & Georgeson, K. E. Hirschsprung disease. Semin. Pediatr. Surg. 17, 266–275 (2008).
Garcia-Barcelo, M.-M. et al. Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung’s disease. Proc. Natl Acad. Sci. USA 106, 2694–2699 (2009).
Jiang, Q. et al. Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to Hirschsprung disease liability. Am. J. Hum. Genet. 96, 581–596 (2015).
Burkardt, D. D., Graham, J. M., Short, S. S. & Frykman, P. K. Advances in Hirschsprung disease genetics and treatment strategies: an update for the primary care pediatrician. Clin. Pediatr. 53, 71−81 (2014).
Lane, C., Milne, E. & Freeth, M. Cognition and behaviour in Sotos syndrome: a systematic review. PLoS ONE 11, e0149189 (2016).
Sio, C. A. et al. Sotos syndrome associated with Hirschsprung’s disease: a new case and exome-sequencing analysis. Pediatr. Res. 82, 87–92 (2017).
Faravelli, F. NSD1 mutations in Sotos syndrome. Am. J. Med. Genet. Part C: Semin. Med. Genet. 137C, 24–31 (2005).
Kurotaki, N. et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 30, 365–366 (2002).
Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein−protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 (2017).
Tam, P. K. Hirschsprung’s disease: a bridge for science and surgery. J. Pediatr. Surg. 51, 18–22 (2016).
Emison, E. S. et al. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am. J. Hum. Genet. 87, 60–74 (2010).
Leventopoulos, G. et al. A clinical study of Sotos Syndrome patients with review of the literature. Pediatr. Neurol. 40, 357–64 (2009).
Baujat, G. & Cormier-Daire, V. Sotos syndrome. Orphanet J. Rare Dis. 2, 36 (2007).
Hirschhorn, J. N. & Daly, M. J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108 (2005).
Orr, N. & Chanock, S. Common genetic variation and human disease. Adv. Genet. 62, 1–32 (2008).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Barlow, A. J., Dixon, J., Dixon, M. & Trainor, P. A. Tcof1 acts as a modifier of Pax3 during enteric nervous system development and in the pathogenesis of colonic aganglionosis. Hum. Mol. Genet. 22, 1206–17 (2013).
Portela, A. & Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 28, 1057–68 (2010).
Choufani, S. et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat. Commun. 6, 10207 (2015).
Hu, N., Strobl-Mazzulla, P. H. & Bronner, M. E. Epigenetic regulation in neural crest development. Dev. Biol. 396, 159–68 (2014).
Torroglosa, A. et al. Epigenetics in ENS development and Hirschsprung disease. Dev. Biol. 417, 209–216 (2016).
Strobl-Mazzulla, P. H., Sauka-Spengler, T. & Bronner-Fraser, M. Histone demethylase JmjD2A regulates neural crest specification. Dev. Cell 19, 460–468 (2010).
Tan, H., Wu, S., Wang, J. & Zhao, Z. K. The JMJD2 members of histone demethylase revisited. Mol. Biol. Rep. 35, 551–556 (2008).
Huang, N. et al. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 17, 3398–3412 (1998).
Sato, Y. & Heuckeroth, R. O. Retinoic acid regulates murine enteric nervous system precursor proliferation, enhances neuronal precursor differentiation, and reduces neurite growth in vitro. Dev. Biol. 320, 185–98 (2008).
Lucio-Eterovic, A. K. et al. Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc. Natl Acad. Sci. USA 107, 16952–16957 (2010).
Goldstein, A. M., Brewer, K. C., Doyle, A. M., Nagy, N. & Roberts, D. J. BMP signaling is necessary for neural crest cell migration and ganglion formation in the enteric nervous system. Mech. Dev. 122, 821–33 (2005).
Ngo, T. et al. Alternative splicing of MXD3 and its regulation of MXD3 levels in glioblastoma. Front. Mol. Biosci. 6, 5 (2019).
Ngo, T., Barisone, G. A., Lam, K. S. & Dίaz, E. MXD3 regulation of DAOY cell proliferation dictated by time course of activation. BMC Cell Biol. 15, 30 (2014).
Evans, P. R., Lee, S. E., Smith, Y. & Hepler, J. R. Postnatal developmental expression of regulator of G protein signaling 14 (RGS14) in the mouse brain. J. Comp. Neurol. 522, 186–203 (2014).
Acknowledgements
We thank all the participants of this research. This work was supported by National Natural Science Foundation of China (31671317, 31471190 and Key Program 81630039), Foundation of Shanghai Municipal Health Commission (201840014) and Shanghai Key Laboratory of Pediatric Gastroenterology and Nutrition (17DZ2272000).
Author information
Authors and Affiliations
Contributions
All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work; W.C. and X.C. designed the research and revised the article; X.-X.Y. drafted the article; X.C. and X.-X.Y. analyzed the data; X.-X.Y., W.-J.W., H.-L.S., Z.-L.W., M.-R.B., Y.J.L., and Y.-M.G. performed the experiment and collected the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent statement
In the present study, written informed consent was obtained from all participants or their parents.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yu, XX., Chu, X., Wu, WJ. et al. Common variation of the NSD1 gene is associated with susceptibility to Hirschsprung’s disease in Chinese Han population. Pediatr Res 89, 694–700 (2021). https://doi.org/10.1038/s41390-020-0933-3
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41390-020-0933-3
This article is cited by
-
Proteomic Analysis Provides a New Sight Into the CRABP1 Expression in the Pathogenesis of Hirschsprung Disease
Biochemical Genetics (2024)
-
Silencing of purinergic receptor P2Y2 inhibited enteric neural crest cell proliferation, invasion and migration via suppressing ERK signaling pathway in Hirschsprung disease
3 Biotech (2023)