Abstract
Background
The S-adenosyl-methionine (SAM) availability is crucial for DNA methylation, an epigenetic mechanism involved in nonsyndromic cleft lip with or without cleft palate (NSCL/P) expression. The aim of this study was to assess the association between single-nucleotide polymorphisms (SNPs) of genes involved in SAM synthesis and NSCL/P in a Chilean population.
Methods
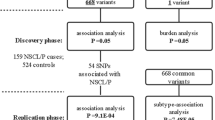
In 234 cases and 309 controls, 18 SNPs in AHCY, MTR, MTRR, and MAT2A were genotyped, and the association between them and the phenotype was evaluated based on additive (allele), dominant, recessive and haplotype models, by odds ratio (OR) computing.
Results
Three deep intronic SNPs of MTR showed a protective effect on NSCL/P expression: rs10925239 (OR 0.68; p = 0.0032; q = 0.0192), rs10925254 (OR 0.66; p = 0.0018; q = 0.0162), and rs3768142 (OR 0.66; p = 0.0015; q = 0.0162). Annotations in expression database demonstrate that the protective allele of the three SNPs is associated with a reduction of MTR expression summed to the prediction by bioinformatic tools of its potentiality to modify splicing sites.
Conclusions
The protective effect against NSCL/P of these intronic MTR SNPs seems to be related to a decrease in MTR enzyme expression, modulating the SAM availability for proper substrate methylation. However, functional analyses are necessary to confirm our findings.
Impact
-
SAM synthesis pathway genetic variants are factors associated to NSCL/P.
-
This article adds new evidence for folate related genes in NSCL/P in Chile.
-
Its impact is to contribute with potential new markers for genetic counseling.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Xavier, M. J., Roman, S. D., Aitken, R. J. & Nixon, B. Transgenerational inheritance: how impacts to the epigenetic and genetic information of parents affect offspring health. Hum. Reprod. Update 25, 518–540 (2019).
Marcho, C., Oluwayiose, O. A. & Pilsner, J. R. The preconception environment and sperm epigenetics. Andrology (2020) (ahead of print).
Slatkin, M. Epigenetic inheritance and the missing heritability problem. Genetics 182, 845–850 (2009).
Trerotola, M., Relli, V., Simeone, P. & Alberti, S. Epigenetic inheritance and the missing heritability. Hum. Genomics 9, 17 (2015).
Nazer, H. J. & Cifuentes, O. L. Prevalence of congenital malformations at birth in Chilean maternity hospitals. Rev. Med. Chil. 142, 1150–1156 (2014).
Saleem, K., Zaib, T., Sun, W. & Fu, S. Assessment of candidate genes and genetic heterogeneity in human non syndromic orofacial clefts specifically non syndromic cleft lip with or without palate. Heliyon 5, e03019 (2019).
Vieira, A. R. Unraveling human cleft lip and palate research. J. Dent. Res. 87, 119–125 (2008).
Wehby, G. L., Pedersen, D. A., Murray, J. C. & Christensen, K. The effects of oral clefts on hospital use throughout the lifespan. BMC Health Serv. Res. 12, 58 (2012). (2012).
Beaty, T. H., Marazita, M. L. & Leslie, E. J. Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Res. 5, 2800 (2016).
Leslie, E. J. et al. Association studies of low-frequency coding variants in nonsyndromic cleft lip with or without cleft palate. Am. J. Med. Genet. A 173, 1531–1538 (2017).
Sharp, G. C. et al. Distinct DNA methylation profiles in subtypes of orofacial cleft. Clin. Epigenet. 9, 63 (2017).
Khan, M. et al. Evaluating LINE-1 methylation in cleft lip tissues and its association with early pregnancy exposures. Epigenomics 10, 105–113 (2018).
Khan, M. et al. LINE-1 methylation in cleft lip tissues: Influence of infant MTHFR c.677C>T genotype. Oral Dis. 25, 1668–1671 (2019).
Xu, Z., Lie, R. T., Wilcox, A. J., Saugstad, O. D. & Taylor, J. A. A comparison of DNA methylation in newborn blood samples from infants with and without orofacial clefts. Clin. Epigenet. 11, 40 (2019).
Fortschegger, K. et al. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol. Cell Biol. 30, 3286–3298 (2010).
Liu, S., Higashihori, N., Yahiro, K. & Moriyama, K. Retinoic acid inhibits histone methyltransferase Whsc1 during palatogenesis. Biochem. Biophys. Res. Commun. 458, 525–530 (2015).
Gou, Y. et al. Protein arginine methyltransferase PRMT1 is essential for palatogenesis. J. Dent. Res. 97, 1510–1518 (2018).
Serefidou, M., Venkatasubramani, A. V. & Imhof, A. The impact of one carbon metabolism on histone methylation. Front. Genet. 10, 764 (2019).
Mato, J. M., Alvarez, L., Ortiz, P. & Pajares, M. A. S-adenosylmethionine synthesis: molecular mechanisms and clinical implications. Pharm. Ther. 73, 265–280 (1997).
Baric, I. et al. S-adenosylhomocysteine hydrolase deficiency in a human: a genetic disorder of methionine metabolism. Proc. Natl Acad. Sci. USA 101, 4234–4239 (2004).
Li, Y. N. et al. Cloning, mapping and RNA analysis of the human methionine synthase gene. Hum. Mol. Genet. 5, 1851–1858 (1996).
Leclerc, D. et al. Cloning and mapping of a cDNA for methionine synthase reductase, a flavoprotein defective in patients with homocystinuria. Proc. Natl Acad. Sci. USA 95, 3059–3064 (1998).
Mao, Z., Liu, S., Cai, J., Huang, Z. Z. & Lu, S. C. Cloning and functional characterization of the 5′-flanking region of human methionine adenosyltransferase 2A gene. Biochem. Biophys. Res. Commun. 248, 479–484 (1998).
Mostowska, A., Hozyasz, K. K. & Jagodzinski, P. P. Maternal MTR genotype contributes to the risk of non-syndromic cleft lip and palate in the Polish population. Clin. Genet. 69, 512–517 (2006).
Blanton, S. H. et al. Folate pathway and nonsyndromic cleft lip and palate. Birth Defects Res. A 91, 50–60 (2011).
Bhaskar, L. V., Murthy, J. & Venkatesh Babu, G. Polymorphisms in genes involved in folate metabolism and orofacial clefts. Arch. Oral Biol. 56, 723–737 (2011).
Murthy, J., Gurramkondab, V. B. & Lakkakula, B. V. Genetic variant in MTRR A66G, but not MTR A2756G, is associated with risk of non-syndromic cleft lip and palate in Indian population. J. Oral Maxillofac. Surg. 27, 782–785 (2015).
Marini, N. J. et al. Sequence variation in folate pathway genes and risks of human cleft lip with or without cleft palate. Am. J. Med. Genet. A 170, 2777–2787 (2016).
Funato, N. & Nakamura, M. Identification of shared and unique gene families associated with oral clefts. Int. J. Oral Sci. 9, 104–109 (2017).
Wang, P. et al. Evidence of interaction between genes in the folate/homocysteine metabolic pathway in controlling risk of non-syndromic oral cleft. Oral Dis. 24, 820–828 (2018).
Marini, N. J., Asrani, K., Yang, W., Rine, J. & Shaw, G. M. Accumulation of rare coding variants in genes implicated in risk of human cleft lip with or without cleft palate. Am. J. Med. Genet. A 179, 1260–1269 (2019).
Jahanbin, A., Shadkam, E., Miri, H. H., Shirazi, A. S. & Abtahi, M. Maternal folic acid supplementation and the risk of oral clefts in offspring. J. Craniofac. Surg. 29, e534–e541 (2018).
Ramírez-Chau, C., Blanco, R., Colombo, A., Pardo, R. & Suazo, J. MTHFR c.677C>T is a risk factor for non-syndromic cleft lip with or without cleft palate in Chile. Oral Dis. 22, 703–708 (2016).
IARC. Common Minimum Technical Standards and Protocols for Biobanks Dedicated to Cancer Research. IARC Technical Publication No. 44 (IARC, 2017).
OECD. OECD Guidelines on Human Biobanks and Genetic Research Databases (Organization for Economic Cooperation and Development (OECD), Paris, 2009).
Anderson, C. A. et al. Data quality control in genetic case–control association studies. Nat. Protoc. 5, 1564–1573 (2010).
Suazo, J. et al. Risk variants in BMP4 promoters for nonsyndromic cleft lip/palate in a Chilean population. BMC Med. Genet. 12, 163 (2011).
Price, A. L., Zaitlen, N. A., Reich, D. & Patterson, N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 11, 459–463 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statistic. Soc. Ser. B 57, 289–300 (1995).
Dudbridge, F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum. Hered. 66, 87–98 (2008).
Desmet, F. O. et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67 (2009).
Lin, H. et al. RegSNPs-intron: a computational framework for predicting pathogenic impact of intronic single nucleotide variants. Genome Biol. 20, 254 (2019).
Salanti, G. et al. Underlying genetic models of inheritance in established type 2 diabetes associations. Am. J. Epidemiol. 170, 537–545 (2009).
Wallenstein, M. B., Shaw, G. M., Yang, W. & Carmichael, S. L. Periconceptional nutrient intakes and risks of orofacial clefts in California. Pediatr. Res. 74, 457–465 (2013).
Salamanca, C. et al. A SHMT1 variant decreases the risk of nonsyndromic cleft lip with or without cleft palate in Chile. Oral Dis. 26, 159–165 (2020).
Liu, N., Zhang, K. & Zhao, H. Haplotype-association analysis. Adv. Genet. 60, 335–405 (2008).
Chorna, L. B., Akopian, H. R., Makukh, H. V. & Fedoryk, I. M. Allelic polymorphism of MTHFR, MTR and MTRR genes in patients with cleft lip and/or palate and their mothers. Tsitol. Genet. 45, 51–56 (2011).
Jin, L. L., Chen, E. J., Hou, W., Liu, X. H. & Hu, Y. The Association between folate pathway genes and cleft lip with or without cleft palate in a Chinese population. Biomed. Environ. Sci. 28, 136–139 (2015).
Wang, W. et al. MTRR, and MTHFR gene polymorphisms and susceptibility to nonsyndromic cleft lip with or without cleft palate. Genet. Test. Mol. Biomark. 20, 297–303 (2016).
Goode, E. L., Potter, J. D., Bigler, J. & Ulrich, C. M. Methionine synthase D919G polymorphism, folate metabolism, and colorectal adenoma risk. Cancer Epidemiol. Biomark. Prev. 13, 157–162 (2004).
Weiner, A. S., Boyarskikh, U. A., Voronina, E. N., Mishukova, O. V. & Filipenko, M. L. Methylenetetrahydrofolate reductase C677T and methionine synthase A2756G polymorphisms influence on leukocyte genomic DNA methylation level. Gene 533, 168–172 (2014).
Finkelstein, J. D. The metabolism of homocysteine: pathways and regulation. Eur. J. Pediatr. 157(Suppl. 2), S40–S44 (1998).
Vaz-Drago, R., Custódio, N. & Carmo-Fonseca, M. Deep intronic mutations and human disease. Hum. Genet. 136, 1093–1111 (2017).
Venables, J. P. Downstream intronic splicing enhancers. FEBS Lett. 581, 4127–4131 (2007).
Cooper, D. N. Functional intronic polymorphisms: buried treasure awaiting discovery within our genes. Hum. Genomics 4, 284–288 (2010).
Abramowicz, A. & Gos, M. Splicing mutations in human genetic disorders: examples, detection, and confirmation. J. Appl. Genet. 59, 253–268 (2018).
Grellscheid, S. N. & Smith, C. W. An apparent pseudo-exon acts both as an alternative exon that leads to nonsense-mediated decay and as a zero-length exon. Mol. Cell. Biol. 26, 2237–2246 (2006).
Santos, J. L. et al. Applicability of the case-parent design in the etiological research of Type 1 diabetes in Chile and other genetically mixed populations. Diabetes Res. Clin. Pract. 43, 143–146 (1999).
Santos, J. L., Pérez, F., Carrasco, E. & Albala, C. Use of case–parents trio for epidemiological studies of association between genetic polymorphisms and complex diseases. Rev. Med. Chil. 130, 1307–1315 (2002).
Acknowledgements
We thank the patients, their families, and healthy individuals who voluntarily cooperated with us, and the staff members of all cleft children rehabilitation centers and blood banks. We also thank the University of Chile BioBank (BTUCH) staff members for their fundamental work. This study was supported by the Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT, Chile) Grant #1170805.
Author information
Authors and Affiliations
Contributions
C.S. made substantial contributions to conception and design, analysis and interpretation of data, drafting, revising, and approved the final version of the manuscript. P.G-H. made substantial contributions to conception, analysis and interpretation of data, drafting, revising, and approved the final version of the manuscript. A.S.R. made substantial contributions to acquisition of data, drafting, revising, and approved the final version of the manuscript. P.A.R. made substantial contributions to acquisition of data, drafting, revising, and approved the final version of the manuscript. R.P. made substantial contributions to the acquisition of data, drafting, revising, and approved the final version of the manuscript. N.L. made substantial contributions to acquisition of data, drafting, revising, and approved the final version of the manuscript. R.P. made substantial contributions to acquisition of data, drafting, revising, and approved the final version of the manuscript. J.S. made substantial contributions to conception and design, acquisition, analysis and interpretation of data, drafting, revising, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
Informed consent for all participants was required, which was approved by the Institutional Review Board of the Faculty of Dentistry, Universidad de Chile (Protocol #2017/07).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Salamanca, C., González-Hormazábal, P., Recabarren, A.S. et al. Genetic variants in S-adenosyl-methionine synthesis pathway and nonsyndromic cleft lip with or without cleft palate in Chile. Pediatr Res 89, 1020–1025 (2021). https://doi.org/10.1038/s41390-020-0994-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41390-020-0994-3
This article is cited by
-
Vitamin B12 Transport Genes and Nonsyndromic Cleft Lip With or Without Cleft Palate in Chile
Reproductive Sciences (2022)


