Abstract
Background
CblC type methylmalonic aciduria (cblC disease) is the most common inborn error of vitamin B12 metabolism and due to mutations in the MMACHC gene. The earlier the diagnosis, the better the prognosis. Therefore, convenient and inexpensive detection method is needed.
Methods
This study selected mutational hot-spot regions in the MMACHC gene which harbors more than 90% of mutant alleles responsible for cblC disease in China. Subsequently, a hot-spot regions multi-PCR Sanger sequencing method (HsRMSS) was designed. The accuracy and efficiency of HsRMSS was validated using samples from 20 cblC families with known MMACHC gene mutations and 50 healthy volunteers. In addition, patients’ clinical phenotypes and molecular genetic features were analyzed.
Results
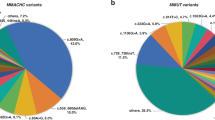
A total of 16 different mutations were identified in 20 cblC families. Among them, the most common mutations were c.609 G>A (26/80, 32.5%), c.567dupT (10/80, 12.5%), c.80A>G (8/80, 10.0%), c.658_660delAAG (8/80, 10.0%) and c.394C>T (6/80, 7.5%), which accounted for over 70% of disease alleles. The HsRMSS results were the same as the results using the whole exon sequencing, with a coincidence rate of 100%.
Conclusion
The HsRMSS targeting the mutational hot-spots of MMACHC gene could be a promising tool to accurately and rapidly diagnose cblC disease in China.
Impact
-
This study reported the development and validation of a hot-spot regions multi-PCR Sanger sequencing method for targeting hotspots which harbor most of the common MMACHC gene mutations reported in Chinese patients with cblC disease.
-
The approach could have a potential clinical application as a rapid diagnosis and screening tool for suspected children with cblC type MMA and population carrier, owing to its high throughput, low cost, and high sensitivity and specificity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 14 print issues and online access
$259.00 per year
only $18.50 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Ling, S. et al. The follow-up of Chinese patients in cblC type methylmalonic acidemia identified through expanded newborn screening. Front. Genet. 13, 805599 (2022).
Arhip, L. et al. Late-onset methylmalonic acidemia and homocysteinemia (cblC disease): systematic review. Orphanet. J. Rare. Dis. 19, 20 (2024).
Xuan, J., Yu, Y., Qing, T., Guo, L. & Shi, L. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 340, 284–295 (2024).
Liu, M. Y. et al. Mutation spectrum of MMACHC in Chinese patients with combined methylmalonic aciduria and homocystinuria. J. Hum. Genet. 55, 621–626 (2010).
Wang, C. et al. Mutation spectrum of MMACHC in Chinese pediatric patients with cobalamin C disease: a case series and literature review. Eur. J. Med. Genet. 62, 103713 (2019).
Ma, X. et al. Clinical features and follow-up study on 55 patients with adolescence-onset methylmalonic acidemia. Zhonghua Er Ke Za Zhi. 62, 520–525 (2024).
Esser, A. J. et al. Versatile enzymology and heterogeneous phenotypes in cobalamin complementation type C disease. iScience 25, 104981 (2022).
Qiao, L. et al. Screening for carriers of pathogenic genes for methylmalonic acidemia and Wilson’s disease in neonates in Qingdao. Zhonghua Er Ke Za Zhi. 58, 596–599 (2020).
Lerner-Ellis, J. P. et al. Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nat. Genet. 38, 93–100 (2006).
He, R. et al. Variable phenotypes and outcomes associated with the MMACHC c.609G>A homologous mutation: long term follow-up in a large cohort of cases. Orphanet. J. Rare Dis. 15, 200 (2020).
Fischer, S. et al. Clinical presentation and outcome in a series of 88 patients with the cblC defect. J. Inherit. Metab. Dis. 37, 831–840 (2014).
Carrillo-Carrasco, N. & Venditti, C. P. Combined methylmalonic acidemia and homocystinuria, cblC type. II. Complications, pathophysiology, and outcomes. J. Inherit. Metab. Dis. 35, 103–114 (2012).
Huemer, M. et al. Phenotype, treatment practice and outcome in the cobalamin-dependent remethylation disorders and MTHFR deficiency: data from the E-HOD registry. J. Inherit. Metab. Dis. 42, 333–352 (2019).
Keller, R. et al. Newborn screening for homocystinurias: recent recommendations versus current practice. J. Inherit. Metab. Dis. 42, 128–139 (2019).
Wang, C. et al. Rapid screening of MMACHC gene mutations by high-resolution melting curve analysis. Mol. Genet. Genom. Med. 8, e1221 (2020).
Yan, Z. Analysis of gene mutations in children with methylmalonic acidemia complicated with homocysteinemia. China Health Care Nutr. 14, 331 (2020).
Wu, S. N., Chen, Q., Chen, Y. X., Liu, F. & Wei, H. Y. Analysis of clinical data of 158 patients with methylmalonic acidemia in Children’s Hospital Affiliated to Zhengzhou University. Chin. J. Pract. Pediatr. 35, 228–232 (2020).
Wang, C. et al. Gene and clinical analysis of neonatal methylmalonic acidemia. J. Chin. Pract. Diagn. Ther. 32, 1181–1183 (2018).
Li, L., Zhang, G. X., Yan, Y. Q., Chen, H. & Han, R. Clinical features and genetic mutation analysis of methylmalonic acidemia patients. J. Shanxi Med. Univ. 51, 986–994 (2020).
Li, L. et al. Genetic variant analysis and prenatal diagnosis for Chinese pedigrees affected with cblC methylmalonic acidemia. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 10, 1058–1064 (2022).
Liu, C., Meng, C. P., Fang, Z. N., Zhao, F. & Wang, Q. H. Screening and mutation analysis of methylmalonic acidemia in newborns in Heze area. Chin. J. Child Health Care. 29, 372–376 (2021).
Chen, C. et al. Analysis of clinical phenotypes and MMACHC gene variants in 65 children with Methylmalonic acidemia and homocysteinemia. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 40, 1086–1092 (2023).
Li, J., Li, Z. & Jin, R. Clinical features and gene analysis of patients with late-onset methylmalonic aciduria and homocystinuria, cobalamin C type. Contemp. Med. 26, 122–124 (2020).
Acknowledgements
We would like to sincerely thank all participants and their families for their cooperation in providing both clinical information and samples for the study.
Funding
The study was supported by the National key Research and Development Program of China (2022YFC2703903) and Rare disease Research Program of E-Town Cooperation & Development Foundation (YCXJ-JZ-2023-017).
Author information
Authors and Affiliations
Contributions
Ping Zheng designed the study, performed the experiments, coordinated and supervised data collection, carried out the data analyses and interpretation, and drafted the manuscript. Chaoji Yu helped perform the experiments, Lina Xie and Xinna Ji provide both clinical information and samples, and critically reviewed and revised the manuscript. Shuo Feng and Yanyan Gao collected data and critically reviewed and revised the manuscript. Xing Wei and Wenli Wu provided the experiment guidance and revised the manuscript. Qian Chen conceived and supervised this study, and revised the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All the samples were collected with informed consent of the individual or his/her parents. The protocols for the study and the written consent were approved by the ethics committee of the Capital Institute of Pediatrics at Beijing, China (Approval ID: SHERLL2023086).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, P., Yu, C., Xie, L. et al. Accelerating the diagnosis of Chinese cblC type MMA patients by multiplex PCR sequencing method. Pediatr Res 98, 950–956 (2025). https://doi.org/10.1038/s41390-025-03841-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41390-025-03841-4
This article is cited by
-
Faster, affordable, and reliable diagnosis of cobalamin C disease by targeted sequencing
Pediatric Research (2025)