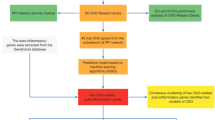
Abstract
Background
Childhood-onset systemic lupus erythematosus (cSLE) exhibits a higher incidence of lupus nephritis (LN) compared to adult-onset SLE (aSLE). However, the underlying molecular mechanisms remain elusive.
Methods
Gene expression datasets from childhood-onset (GSE65391) and adult-onset (GSE72798) LN patients were obtained. Differential gene expression (DEG) and weighted gene co-expression network analysis (WGCNA) identified key modules associated with cLN. Machine learning algorithms (random forest, SVM-RFE) prioritized hub genes, validated by ROC curves. Immune cell profiles analysis (CIBERSORT, ssGSEA, xCell) evaluated immune cell profiles, and functional enrichment (GSEA, GSVA) explored pathway alterations. Molecular docking and drug sensitivity analysis predicted therapeutic targets for otoferlin (OTOF)-high cLN.
Results
Distinct gene expression profiles were revealed between cLN and aLN, with significant enrichment of interferon (IFN) pathways in cLN. OTOF emerged as a high diagnostic biomarker for cLN, and was positively correlated with activated dendritic cells and associated with the TLR signaling pathway in cLN. Molecular docking predicted strong binding of OTOF to JAK inhibitors, supported by CMap analysis showing sensitivity of OTOF-high cLN to these drugs.
Conclusions
IFN-related pathways are crucial for the pathogenesis of cLN. OTOF is a promising blood biomarker and contributes to cLN progression through IFN-related pathways.
Impact
-
The molecular mechanisms by which cSLE is more likely to progress to cLN than aSLE are rarely investigated.
-
The crucial role of interferon pathways in cLN pathogenesis is highlighted.
-
OTOF could serve as an early diagnostic biomarker for cLN and promote cLN progression through TLRs.
-
These insights could guide tailored therapeutic strategies for cLN, addressing a critical gap in current clinical management.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 14 print issues and online access
$259.00 per year
only $18.50 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The public datasets used and analyzed in this study are available from NCBI GEO: GSE65391, GSE72798, GSE99967, and GSE81622.
References
Li, X. Z. et al. Humoral immunity and safety of respiratory virus vaccines in systemic lupus erythematosus population: a meta-analysis based on twenty-five observational studies. Ann. Med. 56, 2392882 (2024).
Katikaneni, D., Morel, L. & Scindia, Y. Animal models of lupus nephritis: the past, present and a future outlook. Autoimmunity 57, 2319203 (2024).
Anders, H. J. et al. Lupus nephritis. Nat. Rev. Dis. Prim. 6, 7 (2020).
Vazzana, K. M. et al. Principles of pediatric lupus nephritis in a prospective contemporary multi-center cohort. Lupus 30, 1660–1670 (2021).
Baqi, N. et al. Lupus nephritis in children: a longitudinal study of prognostic factors and therapy. J. Am. Soc. Nephrol. 7, 924–929 (1996).
Peng, J. et al. Clinical implications of a new DDX58 pathogenic variant that causes lupus nephritis due to RIG-I hyperactivation. J. Am. Soc. Nephrol. 34, 258–272 (2023).
Yoo, E. J. et al. Macrophage transcription factor TonEBP promotes systemic lupus erythematosus and kidney injury via damage-induced signaling pathways. Kidney Int. 104, 163–180 (2023).
Arriens, C. et al. Update on the efficacy and safety profile of voclosporin: an integrated analysis of clinical trials in lupus nephritis. Arthritis Care Res.75, 1399–1408 (2023).
Peyronel, F. et al. Early-onset lupus nephritis. Clin. Kidney J. 17, sfae212 (2024).
Chan, E. Y., Lai, F. F., Ma, A. L. & Chan, T. M. Managing lupus nephritis in children and adolescents. Paediatr. Drugs 26, 145–161 (2024).
Liebermeister, W. et al. Visual account of protein investment in cellular functions. Proc. Natl. Acad. Sci. USA 111, 8488–8493 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics9, 559 (2008).
Sanz, H. et al. SVM-RFE: selection and visualization of the most relevant features through non-linear kernels. BMC Bioinformatics19, 432 (2018).
Rigatti, S. J. Random forest. J. Insur. Med. 47, 31–39 (2017).
Ernst, J. & Bar-Joseph, Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics7, 191 (2006).
Yang, K. et al. CMAP: Complement Map Database. Bioinformatics 29, 1832–1833 (2013).
Hong, Y. et al. Exploring the molecular mechanism of Tripterygium wilfordii Hook F in treating systemic lupus erythematosus via network pharmacology and molecular docking. Clin. Rheumatol. 44, 1549–1569 (2025).
Hong, Y. et al. Environmental triggers and future risk of developing autoimmune diseases: molecular mechanism and network toxicology analysis of bisphenol A. Ecotoxicol. Environ. Saf. 288, 117352 (2024).
Holcar, M., Goropevšek, A. & Avčin, T. Altered homeostasis of regulatory T lymphocytes and differential regulation of STAT1/STAT5 in CD4+ T lymphocytes in childhood-onset systemic lupus erythematosus. J. Rheumatol. 47, 557–566 (2020).
Ding, H. et al. Membrane protein OTOF is a type I interferon-induced entry inhibitor of HIV-1 in macrophages. mBio 13, e0173822 (2022).
Rönnblom, L. & Leonard, D. Interferon pathway in SLE: one key to unlocking the mystery of the disease. Lupus Sci. Med. 6, e000270 (2019).
Parodis, I. et al. Interferon and B-cell signatures inform precision medicine in lupus nephritis. Kidney Int. Rep. 9, 1817–1835 (2024).
Arazi, A. et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat. Immunol. 20, 902–914 (2019).
Ambrose, N. et al. Differences in disease phenotype and severity in SLE across age groups. Lupus 25, 1542–1550 (2016).
Mosca, M. et al. Brief Report: How do patients with newly diagnosed systemic lupus erythematosus present? A multicenter cohort of early systemic lupus erythematosus to inform the development of new classification criteria. Arthritis Rheumatol. 71, 91–98 (2019).
Li, Y., Tang, D., Yin, L. & Dai, Y. New insights for regulatory T cell in lupus nephritis. Autoimmun. Rev. 21, 103134 (2022).
Dmitriev, D. A. et al. Predicting the impact of OTOF gene missense variants on auditory neuropathy spectrum disorder. Int. J. Mol. Sci. 24, 17240 (2023).
Yalcin, E. et al. Evidence that melatonin downregulates Nedd4-1 E3 ligase and its role in cellular survival. Toxicol. Appl. Pharmacol. 379, 114686 (2019).
Zhong, Y. et al. Screening biomarkers for Sjogren’s Syndrome by computer analysis and evaluating the expression correlations with the levels of immune cells. Front. Immunol. 14, 1023248 (2023).
Liu, J., Zhang, X. & Cao, X. Dendritic cells in systemic lupus erythematosus: from pathogenesis to therapeutic applications. J. Autoimmun. 132, 102856 (2022).
Corridoni, D. et al. NOD2 and TLR2 signal via TBK1 and PI31 to direct cross-presentation and CD8 T cell responses. Front. Immunol. 10, 958 (2019).
Swaraj, S. & Tripathi, S. Interference without interferon: interferon-independent induction of interferon-stimulated genes and its role in cellular innate immunity. mBio 15, e0258224 (2024).
Oikonomou, V. et al. The role of interferon-γ in autoimmune polyendocrine syndrome type 1. N. Engl. J. Med. 390, 1873–1884 (2024).
Kandhaya-Pillai, R. et al. TNF-α/IFN-γ synergy amplifies senescence-associated inflammation and SARS-CoV-2 receptor expression via hyper-activated JAK/STAT1. Aging Cell 21, e13646 (2022).
Acknowledgements
This study was a re-analysis based on published data from the GEO database. We would like to thank these databases for sharing the data.
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province (grant number LTGY23H100001), Science and Technology Plan Project of Wenzhou Municipality, China (grant numbers Y20220389, Y20220045), National innovation and Entrepreneurship Training Program for College Students (grant number 202310343025); Zhejiang College Students Innovative Entrepreneurial Training Program (New Young Talent Program) (grant number 2023R413025).
Author information
Authors and Affiliations
Contributions
Xinlei Wu designed the research, analyzed the data, and wrote the manuscript; Xinyue Liang and Yang Li analyzed and interpreted the data; Siyan Chen and Yuanyua Xie corrected the manuscript. Chunyan Hua and Sheng Gao designed this study, revised the manuscript, and received funding support.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, X., Liang, X., Li, Y. et al. Identification of childhood-onset lupus nephritis through integrative bioinformatics: otoferlin as a novel biomarker. Pediatr Res (2025). https://doi.org/10.1038/s41390-025-04171-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41390-025-04171-1