Abstract
Background
Aberrant Wnt signaling has been implicated in prostate cancer tumorigenesis and metastasis in preclinical models but the impact of genetic alterations in Wnt signaling genes in men with advanced prostate cancer is unknown.
Methods
We utilized the Prostate Cancer Precision Medicine Multi-Institutional Collaborative Effort (PROMISE) clinical-genomic database for this retrospective analysis. Patients with activating mutations in CTNNB1 or RSPO2 or inactivating mutations in APC, RNF43, or ZNRF3 were defined as Wnt-altered, while those lacking such alterations were defined as Wnt non-altered. We compared patient characteristics and clinical outcomes as well as co-occurring genetic alterations according to Wnt alteration status.
Results
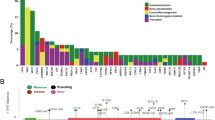
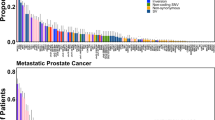
Of the 1498 patients included, 193 (12.9%) were Wnt-altered. These men had a statistically significant 2-fold increased prevalence of liver and lung metastases as compared with Wnt non-altered patients at the time of initial diagnosis, (4.66% v 2.15% ; 6.22% v 3.07%), first metastatic disease diagnosis (10.88% v 5.29%; 13.99% v 6.21%), and CRPC development (11.40% v 6.36%; 12.95% v 5.29%). Wnt alterations were associated with more co-occurring alterations in RB1 (10.4% v 6.2%), AR (38.9% vs 25.7%), SPOP (13.5% vs 4.1%), FOXA1 (6.7% vs 2.8%), and PIK3CA (10.9% vs 5.1%). We found no significant differences in overall survival or other clinical outcomes from initial diagnosis, first metastatic disease, diagnosis of CRPC, or with AR inhibition for mCRPC between the Wnt groups.
Conclusions
Wnt-altered patients with prostate cancer have a higher prevalence of visceral metastases and are enriched in RB1, AR, SPOP, FOXA1, and PIK3CA alterations. Despite these associations, Wnt alterations were not associated with worse survival or treatment outcomes in men with advanced prostate cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request and per PROMISE consortium guidelines.
References
Siegel R, Miller K, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30.
Mohler JL, Antonarakis ES, Armstrong AJ, D’Amico AV, Davis BJ, Dorff T, et al. Prostate Cancer, Version 2.2019, NCCN Clinical Practice guidelines in Oncology. J Natl Compr Canc Netw. 2019;17:479–505.
Dehm SM, Tindall DJ. Androgen receptor structural and functional elements: role and regulation in prostate cancer. Mol Endocrinol. 2007;21:2855–63.
Mohler JL, Gregory CW, Ford OH, Kim D, Weaver CM, Petrusz P, et al. The androgen axis in recurrent prostate cancer. Clin Cancer Res. 2004;10:440–8.
Zhao XY, Malloy PJ, Krishnan AV, Swami S, Navone NM, Peehl DM, et al. Glucocorticoids can promote androgen-independent growth of prostate cancer cells through a mutated androgen receptor. Nat Med. 2000;6:703–6.
Debes JD, Tindall DJ. Mechanisms of androgen refractory prostate cancer. N Engl J Med. 2004;351:1488–90.
Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–98.
Leibold J, Ruscetti M, Cao Z, Ho YJ, Baslan T, Zou M, et al. Somatic Tissue Engineering in Mouse Models Reveals an Actionable Role for WNT Pathway Alterations in Prostate Cancer Metastasis. Cancer Discov. 2020;10:1038–57.
Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF. WNT/β-catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin Cancer Res. 2019;25:3074–83. https://doi.org/10.1158/1078-0432.CCR-18-1942.
Croce JC, McClay DR. Evolution of the Wnt pathways. Methods Mol Biol. 2008;469:3.
Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–99.
Yang F, Li X, Sharma M, Sasaki CY, Longo DL, Lim B, et al. Linking beta-catenin to androgen-signaling pathway. J Biol Chem. 2002;277:11336–44.
Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28.
Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63:920–6.
Jung SJ, Oh S, Lee GT, Chung J, Min K, Yoon J, et al. Clinical significance of Wnt/β-catenin signalling and androgen receptor expression in prostate cancer. World J Mens Health. 2013;31:36–46.
Huang SP, Ting WC, Chen LM, Huang LC, Liu CC, Chen CW, et al. Association analysis of Wnt pathway genes on prostate-specific antigen recurrence after radical prostatectomy. Ann Surg Oncol. 2010;17:312–22.
Velho PI, Fu W, Wang H, Mirkheshti N, Qazi F, Lima FAS, et al. Wnt-pathway activating mutations are associated with resistance to first-line abiraterone and enzalutamide in castration-resistant prostate cancer. Eur Urol. 2020;77:14–21.
Gupta S, Halabi S, Kemeny G, Anand M, Giannakakou P, Nanus DM, et al. Circulating Tumor Cell Genomic Evolution and Hormone Therapy Outcomes in Men with Metastatic Castration-Resistant Prostate Cancer. Mol Cancer Res. 2021;19:1040–50.
Koshkin VS, Patel VG, Ali A, Bilen MA, Ravindranathan D, Park JJ, et al. PROMISE: a real-world clinical-genomic database to address knowledge gaps in prostate cancer. Prostate Cancer Prostatic Dis. 2022;25:388–96. https://doi.org/10.1038/s41391-021-00433-1.
Ritch E, Fu SYF, Herberts C, Wang G, Warner EW, Schönlau E, et al. Identification of Hypermutation and Defective Mismatch Repair in ctDNA from Metastatic Prostate Cancer. Clin Cancer Res. 2020;26:1114–25. https://doi.org/10.1158/1078-0432.CCR-19-1623.
Tu J, Park S, Yu W, Zhang S, Wu L, Carmon K, et al. The most common RNF43 mutant G659Vfs*41 is fully functional in inhibiting Wnt signaling and unlikely to play a role in tumorigenesis. Sci Rep. 2019;9:18557. https://doi.org/10.1038/s41598-019-54931-3.
Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov. 2018;8:444–57. https://doi.org/10.1158/2159-8290.CD-17-0937.
Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–6. https://doi.org/10.1126/science.aab0917.
McKay RR, McGrath JE, Pan E, Farrell A, Beltran H, Heath E, et al. Characterization and impact of canonical Wnt Signaling Pathway (WSP) alterations on outcomes of metastatic prostate cancer. JCO. 2022;40:5053–5053. https://doi.org/10.1200/JCO.2022.40.16_suppl.5053.
Halabi S, Yang Q, Roy A, Luo B, Araujo JC, Logothetis C, et al. External Validation of a Prognostic Model of Overall Survival in Men With Chemotherapy-Naïve Metastatic Castration-Resistant Prostate Cancer. J Clin Oncol. 2023;41:2736–46. https://doi.org/10.1200/JCO.22.02661.
Halabi S, Kelly WK, Ma H, Zhou H, Solomon NC, Fizazi K, et al. Meta-analysis evaluating the impact of site of metastasis on overall survival in men with castration-resistant prostate cancer. J Clin Oncol. 2016;34:1652–9.
Baciarello G, Özgüroğlu M, Mundle S, Leitz G, Richarz U, Hu P, et al. Impact of abiraterone acetate plus prednisone in patients with castration-sensitive prostate cancer and visceral metastases over four years of follow-up: A post-hoc exploratory analysis of the LATITUDE study. Eur J Cancer. 2022;162:56–64. https://doi.org/10.1016/j.ejca.2021.11.026.
Astarita EM, Maloney SM, Hoover CA, Berkeley BJ, VanKlompenberg MK, Nair TM, et al. Adenomatous Polyposis Coli loss controls cell cycle regulators and response to paclitaxel in MDA-MB-157 metaplastic breast cancer cells. PLoS One. 2021;16:e0255738. https://doi.org/10.1371/journal.pone.0255738.
Tighe A, Johnson VL, Taylor SS. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci. 2004;117:6339–53. https://doi.org/10.1242/jcs.01556.
Stangl A, Wilner C, Li P, Maahs L, Hwang C, Pilling A. Molecular features and race-associated outcomes of SPOP-mutant metastatic castration-resistant prostate cancer. Prostate. 2023;83:524–33. https://doi.org/10.1002/pros.24481.
Nakazawa M, Fang M, H Marshall C, Lotan TL, Isaacsson Velho P, Antonarakis ES. Clinical and genomic features of SPOP-mutant prostate cancer. Prostate. 2022;82:260–8. https://doi.org/10.1002/pros.24269.
Swami U, Graf RP, Nussenzveig RH, Fisher V, Tukachinsky H, Schrock AB, et al. SPOP Mutations as a Predictive Biomarker for Androgen Receptor Axis-Targeted Therapy in De Novo Metastatic Castration-Sensitive Prostate Cancer. Clin Cancer Res. 2022;28:4917–25. https://doi.org/10.1158/1078-0432.CCR-22-2228.
Parolia A, Cieslik M, Chu SC, Xiao L, Ouchi T, Zhang Y, et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature. 2019;571:413–8. https://doi.org/10.1038/s41586-019-1347-4.
Li S, Lavrijsen M, Bakker A, Magierowski M, Magierowska K, Liu P, et al. Commonly observed RNF43 mutations retain functionality in attenuating Wnt/β-catenin signaling and unlikely confer Wnt-dependency onto colorectal cancers. Oncogene. 2020;39:3458–72. https://doi.org/10.1038/s41388-020-1232-5.
Schweizer MT, Antonarakis ES, Bismar TA, Guedes LB, Cheng HH, Tretiakova MS, et al. Genomic Characterization of Prostatic Ductal Adenocarcinoma Identifies a High Prevalence of DNA Repair Gene Mutations. JCO Precis Oncol. 2019;3:PO.18.00327 https://doi.org/10.1200/PO.18.00327.
Gillard M, Lack J, Pontier A, Gandla D, Hatcher D, Sowalsky AG, et al. Integrative Genomic Analysis of Coincident Cancer Foci Implicates CTNNB1 and PTEN Alterations in Ductal Prostate Cancer. Eur Urol Focus. 2019;5:433–42. https://doi.org/10.1016/j.euf.2017.12.003.
Author information
Authors and Affiliations
Contributions
Concept and design: Andrew J. Armstrong, Rana R. McKay, Elizabeth Pan, Amanda Broderick. Acquisition, analysis, or interpretation of data: All authors. Drafting of the manuscript: Andrew J. Armstrong, Rana R. McKay, Elizabeth Pan, Amanda Broderick, Alec Chu, Jinju Li. Critical review of the manuscript for important intellectual content: All authors. Statistical analysis: Jinju Li, Alec Chu.
Corresponding authors
Ethics declarations
Competing interests
AJA has received support (to Duke) from the NIH/NCI, PCF/Movember, DOD, Astellas, Pfizer, Bayer, Janssen, Dendreon, BMS, AstraZeneca, Merck, Forma, Celgene, Amgen, Novartis and has consulting or advising relationships with Astellas, Pfizer, Bayer, Janssen, BMS, AstraZeneca, Merck, Forma, Celgene, Myovant, Exelixis, GoodRx, Novartis, Medscape, MJH, Z Alpha, Telix outside the submitted work. CH reported owning stock in Johnson and Johnson; receiving personal fees from Tempus, Genzyme, EMD Sorono, OncLive/MJH Life Sciences, Merck, and grants from Merck, Bausch Health, Genentech, Bayer, and AstraZeneca outside the submitted work. TBD reported consulting/advisory relationships with AstraZeneca, Bayer, Janssen, Sanofi. DK reported consulting/Honoraria for Exelixis, Pfizer. MJH life science; Advisory board for Eisai, Exelixis; institutional funding from Genetech, Exelixis; and speakers bureau for Aveo, Jannsen, Astellas. VSK reported consulting or advisory role for AstraZeneca, Janssen, Pfizer, EMD Serono, Seagen, Astellas, Guidepoint, GLG and ExpertConnect; has received research funding for the institution from Endocyte/Novartis, Nektar, Gilead, Janssen, Taiho, Merck and Seagen, individual research funding from Eli Lilly and is supported by the Prostate Cancer Foundation. MTS reported being a consultant and/or received Honoria from Sanofi, AstraZeneca, Janssen, Fibrogen, and Pfizer. He has received research funding to his institution from Novartis, Zenith Epigenetics, Eli Lilly, Bristol Myers Squibb, Merck, Immunomedics, Janssen, AstraZeneca, Pfizer, Hoffmann-La Roche, Tmunity, SignalOne Bio, Epigenetix, Xencor, Incyte and Ambrx, Inc. RRM reported consulting or advisory board for Ambrx, AstraZeneca, Aveo, Bayer, Bristol-Myers Squibb, Calithera, Caris, Dendreon, Exelixis, Eisai, Johnson & Johnson, Lilly, Merck, Myovant, Novartis, Pfizer, Sanofi, Seagen, Sorrento, Telix, Tempus. She has received istitutional research support Exelixis, BMS, AstraZeneca, Oncternal, Artera, Tempus. RG has received institutional research funding from Endocyte/Advanced Accelerator Applications, Pfizer, Amgen, Immunomedics, and Xencor outside the submitted work. MAB reported receiving personal fees from Exelixis, Bayer, Bristol Myers Squibb, Eisai, Pfizer, AstraZeneca, Janssen, Calithera Biosciences, Genomic Health, Nektar, EMD Serono, SeaGen, and Sanofi and grants from Merck, Xencor, Bayer, Bristol Myers Squibb, Genentech/Roche, SeaGen, Incyte, Nektar, AstraZeneca, Tricon Pharmaceuticals, Genome & Company, AAA, Peloton Therapeutics, and Pfizer outside the submitted work. AT reported receiving personal fees from Genzyme, Exelixis, Seattle Genetics, Deka Biosciences, and Aldi Biosciences and grants from EMD Serono and Aravive outside the submitted work. PCB reported receiving personal fees from AVEO Oncology, Bayer, Pfizer, Caris Life Sciences, AstraZeneca, Myovant, Curio Science, Targeted Oncology, Bristol Myers Squibb, MJH Life Sciences, Seagen, and Vaniam Group and grants from EMD Serono outside the submitted work ASA reported serving as an advisor to AstraZeneca, Bristol-Myers Squibb, Merck, and Pfizer. He has previously received institutional research funding from Arcus Biosciences, Astellas, AstraZeneca, Bayer, Bristol-Myers Squibb, Celgene, Genentech, Merck Sharp & Dohme, Janssen, Mirati Therapeutics, Progenics, Prometheus Laboratories, and Roche. He has also received travel and accommodation from Bristol-Myers Squibb and Merck.
Ethics approval and consent to participate
Not applicable. This was deemed IRB and patient consent exempt as a retrospective and centrally deidentified study with a central database.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Broderick, A., Pan, E., Li, J. et al. Clinical implications of Wnt pathway genetic alterations in men with advanced prostate cancer. Prostate Cancer Prostatic Dis 28, 419–426 (2025). https://doi.org/10.1038/s41391-024-00869-1
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41391-024-00869-1