Fig. 1
From: Clinical and translational values of spatial transcriptomics
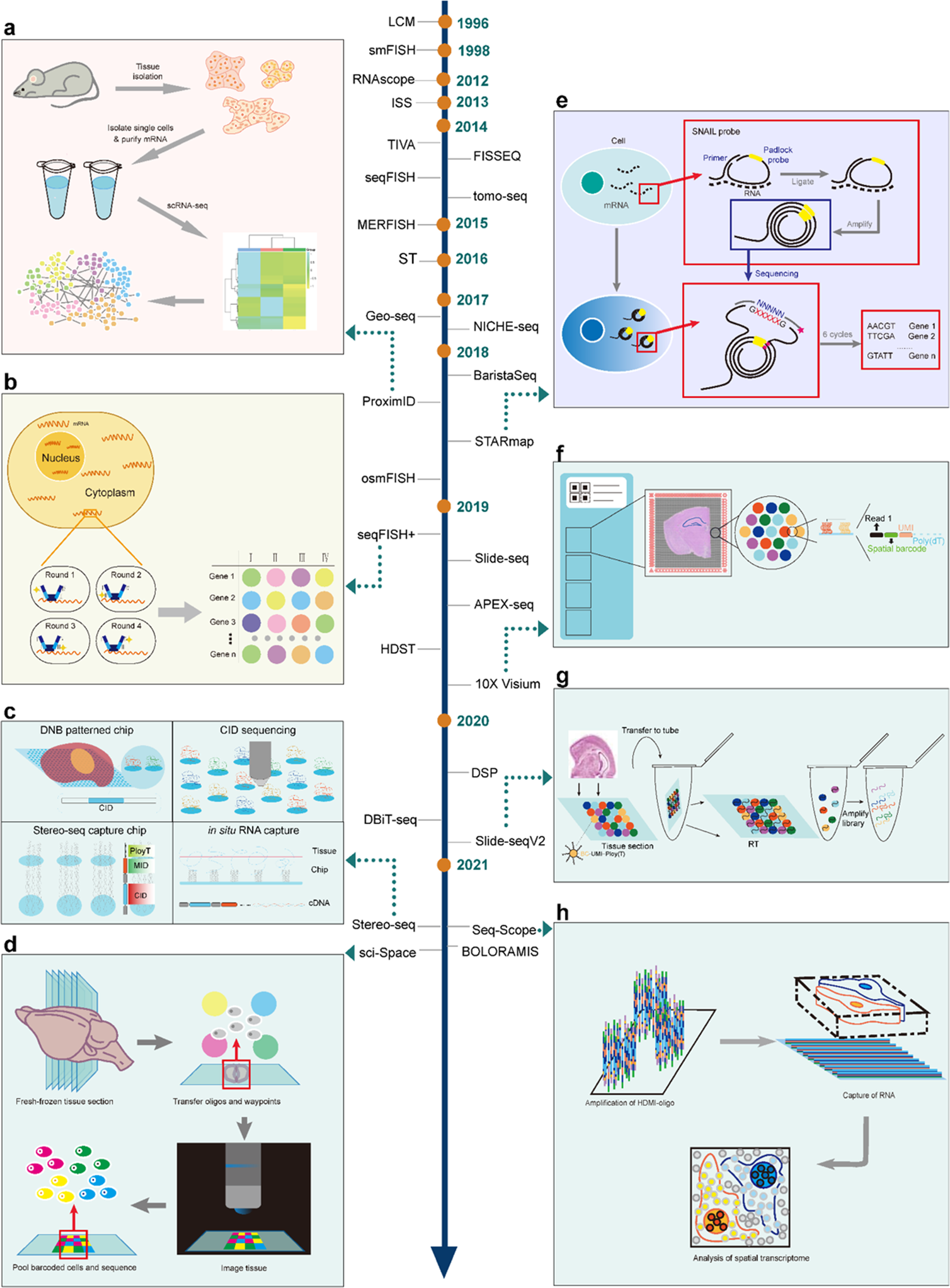
Development of spatial transcriptomic technologies. Representative technologies were exhibited with detailed schematic diagram, including ProximID, STARmap, seqFISH+, 10× Visium, Slide-seqV2, Stereo-seq, Seq-Scope and sci-Space. The development of ST technologies was shown in the middle part with method names and years. a The principle from cell isolation to interaction networks for ProximID; b The principle of repeated in situ hybridizations for seqFISH+; c The principle of Stereo-seq; d The principle of sci-Space from fresh-frozen sectioning, oligos and waypoints transferring, and pooled barcoded cell positioning and sequencing, to imaging and reading; e The principle of in situ mRNA preparation, SNAIL probe function, and in situ sequencing for STARmap; f The principle of in situ capturing from tissue grids to spot selection, from sample setting to quality control, and from partial reads to spatial barcodes for 10× Visium; g The principle of Slide-seqV2 from tissue coating to library amplification; h The principle of Seq-Scope from high-definition map coordinate identifier (HDMI)-oligo amplification to RNA capture from frozen section to achieve spatial transcriptome analysis at the single cell levels
