Abstract
Study design
An integrated bioinformatics data study.
Objectives
This study uses bioinformatics analysis to map the microglial landscape, investigate key signaling pathways, and reveal the molecular mechanisms that facilitate SCI recovery.
Setting
Beijing Tsinghua Changgung Hospital, School of Clinical Medicine, Tsinghua University.
Methods
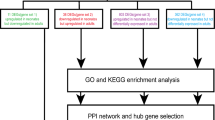
In this study, we performed an integrative bioinformatics analysis of single-cell RNA sequencing (scRNA-seq), spatial transcriptomic (ST), and bulk RNA-seq datasets from the Gene Expression Omnibus (GEO), utilizing R packages (Seurat, DESeq2, limma, GSVA) and the Enrichr platform.
Results
Single-cell and spatial transcriptomic profiling uncovered dynamic shifts in the microglial landscape post-SCI, characterized by the suppression of innate microglial populations alongside the expansion of reactive microglial subsets. Mechanistically, the TGFβ signaling pathway was identified as a critical regulator of innate microglial migration, promoting functional recovery after SCI. Conversely, reactive microglia exhibiting heightened Trem2 expression were found to exacerbate neuroinflammatory responses and drive neural cell death.
Conclusions
These findings collectively indicate that targeting the dual regulatory axis of Trem2-mediated neuroinflammation and TGFβ-driven repair mechanisms may offer a synergistic therapeutic strategy to enhance functional recovery following spinal cord injury.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data sources and handling are described in the Materials and Methods. The source code is proprietary but available upon request for non-commercial use by contacting Yifeng Sun at syf498054232@163.com.
References
Tian T, Zhang S, Yang M. Recent progress and challenges in the treatment of spinal cord injury. Protein Cell. 2023;14:635–52.
Ding W, Hu S, Wang P, Kang H, Peng R, Dong Y, et al. Spinal cord injury: the global incidence, prevalence, and disability from the global burden of disease study 2019. Spine (Phila Pa 1976). 2022;47:1532–40.
Orr MB, Gensel JC. Spinal cord injury scarring and inflammation: therapies targeting glial and inflammatory responses. Neurotherapeutics. 2018;15:541–53.
Liu W, Tang Y, Feng J. Cross talk between activation of microglia and astrocytes in pathological conditions in the central nervous system. Life Sci. 2011;89:141–6.
Utz SG, See P, Mildenberger W, Thion MS, Silvin A, Lutz M, et al. Early fate defines microglia and non-parenchymal brain macrophage development. Cell. 2020;181:557–73.e518.
Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–8.
Kurpius D, Nolley EP, Dailey ME. Purines induce directed migration and rapid homing of microglia to injured pyramidal neurons in developing hippocampus. Glia. 2007;55:873–84.
Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–7.
Mesquida-Veny F, Del Río JA, Hervera A. Macrophagic and microglial complexity after neuronal injury. Prog Neurobiol. 2021;200:101970.
Yeh FL, Hansen DV, Sheng M. TREM2, microglia, and neurodegenerative diseases. Trends Mol Med. 2017;23:512–33.
Jiang C, Chen Z, Wang X, Zhang Y, Guo X, Fan H, et al. Curcumin-activated olfactory ensheathing cells improve functional recovery after spinal cord injury by modulating microglia polarization through APOE/TREM2/NF-κB signaling pathway. J Neuroimmune Pharmacol. 2023;18:476–94.
David S, Kroner A. Repertoire of microglial and macrophage responses after spinal cord injury. Nat Rev Neurosci. 2011;12:388–99.
Dai M, Pei X, Wang X-J. Accurate and fast cell marker gene identification with COSG. Brief Bioinform. 2022;23:bbab579.
Sun Y, Zhang C, Fang Q, Zhang W, Liu W. Abnormal signal pathways and tumor heterogeneity in osteosarcoma. J Transl Med. 2023;21:99.
Hou J, Bi H, Ge Q, Teng H, Wan G, Yu B, et al. Heterogeneity analysis of astrocytes following spinal cord injury at single-cell resolution. FASEB J. 2022;36:e22442.
Li Y, He X, Kawaguchi R, Zhang Y, Wang Q, Monavarfeshani A, et al. Microglia-organized scar-free spinal cord repair in neonatal mice. Nature. 2020;587:613–8.
Wang R, Zhou R, Chen Z, Gao S, Zhou F. The glial cells respond to spinal cord injury. Front Neurol. 2022;13:844497.
Sierra A, Encinas JM, Deudero JJ, Chancey JH, Enikolopov G, Overstreet-Wadiche LS, et al. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell. 2010;7:483–95.
Weldon DT, Rogers SD, Ghilardi JR, Finke MP, Cleary JP, O’Hare E, et al. Fibrillar β-amyloid induces microglial phagocytosis, expression of inducible nitric oxide synthase, and loss of a select population of neurons in the rat CNS in vivo. J Neurosci. 1998;18:2161–73.
Fu R, Shen Q, Xu P, Luo JJ, Tang Y. Phagocytosis of microglia in the central nervous system diseases. Mol Neurobiol. 2014;49:1422–34.
Dibaj P, Nadrigny F, Steffens H, Scheller A, Hirrlinger J, Schomburg ED, et al. NO mediates microglial response to acute spinal cord injury under ATP control in vivo. Glia. 2010;58:1133–44.
Kettenmann H, Hanisch U-K, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev. 2011;91:461–553.
Saber M, Kokiko-Cochran O, Puntambekar SS, Lathia JD, Lamb BT. Triggering receptor expressed on myeloid cells 2 deficiency alters acute macrophage distribution and improves recovery after traumatic brain injury. J Neurotrauma. 2017;34:423–35.
Kobayashi M, Konishi H, Sayo A, Takai T, Kiyama H. TREM2/DAP12 signal elicits proinflammatory response in microglia and exacerbates neuropathic pain. J Neurosci. 2016;36:11138–50.
Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, et al. Identification of a unique TGF-β–dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–43.
Lodge PA, Sriram S. Regulation of microglial activation by TGF-β, IL-10, and CSF-1. J Leukoc Biol. 1996;60:502–8.
Baria MR, Miller MM, Burner T, Hake T, Kim D, Magnussen R, et al. Platelet-rich plasma content of active spinal cord injured patients: a controlled laboratory study. Am J Phys Med Rehabil. 2021;100:651–5.
McTigue DM, Popovich PG, Morgan TE, Stokes BT. Localization of transforming growth factor-beta1 and receptor mRNA after experimental spinal cord injury. Exp Neurol. 2000;163:220–30.
Kaiser J, Maibach M, Piovesana E, Salpeter I, Escher N, Ormen Y, et al. TGFβ1 induces axonal outgrowth via ALK5/PKA/SMURF1-mediated degradation of RhoA and stabilization of PAR6. eNeuro. 2020;7:ENEURO.0104-20.2020.
Gao H, Di J, Clausen BH, Wang N, Zhu X, Zhao T, et al. Distinct myeloid population phenotypes dependent on TREM2 expression levels shape the pathology of traumatic versus demyelinating CNS disorders. Cell Rep. 2023;42:112629.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81702667). Beijing Tsinghua Changgung Hospital Fund (Grant No. 202510S004), China Postdoctoral Science Foundation (Grant No.8206300728).
Author information
Authors and Affiliations
Contributions
YS conceived the study and generated figures and wrote the manuscript, QZ collected and analysed the data, QF performed part of the data analysis and was responsible for manuscript revision, JL performed a part of data analysis. CZ and WL help to generated Figures. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This is a bioinformatic analysis; therefore, ethics approval and consent to participate are not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, Q., Liu, J., Fang, Q. et al. Microglial landscape and signaling in spinal cord injury. Spinal Cord 63, 418–425 (2025). https://doi.org/10.1038/s41393-025-01103-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41393-025-01103-y