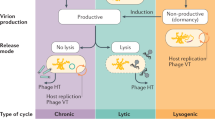
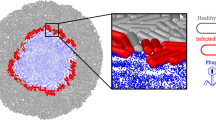
Abstract
Since their discovery, bacteriophages have been traditionally regarded as the natural enemies of bacteria. However, recent advances in molecular biology techniques, especially data from “omics” analyses, have revealed that the interplay between bacterial viruses and their hosts is far more intricate than initially thought. On the one hand, we have become more aware of the impact of viral predation on the composition and genetic makeup of microbial communities thanks to genomic and metagenomic approaches. Moreover, data obtained from transcriptomic, proteomic, and metabolomic studies have shown that responses to phage predation are complex and diverse, varying greatly depending on the bacterial host, phage, and multiplicity of infection. Interestingly, phage exposure may alter different phenotypes, including virulence and biofilm formation. The complexity of the interactions between microbes and their viral predators is also evidenced by the link between quorum-sensing signaling pathways and bacteriophage resistance. Overall, new data increasingly suggests that both temperate and virulent phages have a positive effect on the evolution and adaptation of microbial populations. From this perspective, further research is still necessary to fully understand the interactions between phage and host under conditions that allow co-existence of both populations, reflecting more accurately the dynamics in natural microbial communities.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
d’Herelle F. Sur un microbe invisible antagoniste des bacilles dysentériques. CR Acad Sci. 1917;165:373–75.
Twort FW. An investigation on the nature of ultra-microscopic viruses. Lancet. 1915;ii:1241–43.
Abedon ST, García P, Mullany P, Aminov R. Editorial: phage therapy: past, present and future. Front Microbiol. 2017;8:981.
Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1:31–45.
Rascovan N, Duraisamy R, Desnues C. Metagenomics and the human virome in asymptomatic individuals. Annu Rev Microbiol. 2016;70:125–41.
Salmond GP, Fineran PC. A century of the phage: past, present and future. Nat Rev Microbiol. 2015;13:777–86.
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21.
Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300.
Filippov AA, Sergueev KV, He Y, Huang XZ, Gnade BT, Mueller AJ, et al. Bacteriophage-resistant mutants in Yersinia pestis: identification of phage receptors and attenuation for mice. PLoS ONE. 2011;6:e25486.
Scott AE, Timms AR, Connerton PL, Loc Carrillo C, Adzfa Radzum K, Connerton IF. Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 2007;3:e119.
Hosseinidoust Z, van de Ven TG, Tufenkji N. Evolution of Pseudomonas aeruginosa virulence as a result of phage predation. Appl Environ Microbiol. 2013;79:6110–16.
Davies EV, James CE, Williams D, O’Brien S, Fothergill JL, Haldenby S, et al. Temperate phages both mediate and drive adaptive evolution in pathogen biofilms. Proc Natl Acad Sci USA. 2016;113:8266–71.
Cairns J, Frickel J, Jalasvuori M, Hiltunen T, Becks L. Genomic evolution of bacterial populations under coselection by antibiotics and phage. Mol Ecol. 2016;26:1848–59.
Koskella B, Brockhurst MA. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 2014;38:916–31.
Fortier LC, Sekulovic O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013;4:354–65.
Haaber J, Leisner JJ, Cohn MT, Catalan-Moreno A, Nielsen JB, Westh H, et al. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat Commun. 2016;7:13333.
Modi SR, Lee HH, Spina CS, Collins JJ. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499:219–22.
Keen EC, Bliskovsky VV, Malagon F, Baker JD, Prince JS, Klaus JS, et al. Novel “superspreader” bacteriophages promote horizontal gene transfer by transformation. mBio. 2017;8:e02115–16.
Poranen MM, Ravantti JJ, Grahn AM, Gupta R, Auvinen P, Bamford DH. Global changes in cellular gene expression during bacteriophage PRD1 infection. J Virol. 2006;80:8081–88.
Ainsworth S, Zomer A, Mahony J, van Sinderen D. Lytic infection of Lactococcus lactis by bacteriophages Tuc2009 and c2 triggers alternative transcriptional host responses. Appl Environ Microbiol. 2013;79:4786–98.
Doron S, Fedida A, Hernández-Prieto MA, Sabehi G, Karunker I, Stazic D, et al. Transcriptome dynamics of a broad host-range cyanophage and its hosts. ISME J. 2016;10:1437–55.
De Smet J, Zimmermann M, Kogadeeva M, Ceyssens PJ, Vermaelen W, Blasdel B, et al. High coverage metabolomics analysis reveals phage-specific alterations to Pseudomonas aeruginosa physiology during infection. ISME J. 2016;10:1823–35.
Blasdel BG, Chevallereau A, Monot M, Lavigne R, Debarbieux L. Comparative transcriptomics analyses reveal the conservation of an ancestral infectious strategy in two bacteriophage genera. ISME J. 2017;11:1988–96.
Fernández L, González S, Campelo AB, Martínez B, Rodríguez A, García P. Low-level predation by lytic phage phiIPLA-RODI promotes biofilm formation and triggers the stringent response in Staphylococcus aureus. Sci Rep. 2017;7:40965.
Lavigne R, Lecoutere E, Wagemans J, Cenens W, Aertsen A, Schoofs L, et al. A multifaceted study of Pseudomonas aeruginosa shutdown by virulent podovirus LUZ19. mBio. 2013;24:e00061–13.
Osterhout RE, Figueroa IA, Keasling JD, Arkin AP. Global analysis of host response to induction of a latent bacteriophage. BMC Microbiol. 2007;7:82.
Ravantti JJ, Ruokoranta TM, Alapuranen AM, Bamford DH. Global transcriptional responses of Pseudomonas aeruginosa to phage PRR1 infection. J Virol. 2008;82:2324–29.
Veses-Garcia M, Liu X, Rigden DJ, Kenny JG, McCarthy AJ, Allison HE. Transcriptomic analysis of Shiga-toxigenic bacteriophage carriage reveals a profound regulatory effect on acid resistance in Escherichia coli. Appl Environ Microbiol. 2015;81:8118–25.
Zhao X, Chen C, Shen W, Huang G, Le S, Lu S, et al. Global transcriptomic analysis of interactions between Pseudomonas aeruginosa and bacteriophage PaP3. Sci Rep. 2016;6:19237.
Fallico V, Ross RP, Fitzgerald GF, McAuliffe O. Genetic response to bacteriophage infection in Lactococcus lactis reveals a four-strand approach involving induction of membrane stress proteins, d-alanylation of the cell wall, maintenance of proton motive force, and energy conservation. J Virol. 2011;85:12032–42.
Dendooven T, Van den Bossche A, Hendrix H, Ceyssens PJ, Voet M, Bandyra KJ, et al. Viral interference of the bacterial RNA metabolism machinery. RNA Biol. 2017;14:6–10.
Van den Bossche A, Hardwick SW, Ceyssens PJ, Hendrix H, Voet M, Dendooven T, et al. Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome. Elife. 2016;5:e16413.
Chevallereau A, Blasdel BG, De Smet J, Monot M, Zimmermann M, Kogadeeva M, et al. Next-Generation “-omics” approaches reveal a massive alteration of host RNA metabolism during bacteriophage infection of Pseudomonas aeruginosa. PLoS Genet. 2016;12:e1006134.
Aranda M, Maule A. Virus-induced host gene shutoff in animals and plants. Virology. 1998;243:261–67.
Campoy S, Hervàs A, Busquets N, Erill I, Teixidó L, Barbé J. Induction of the SOS response by bacteriophage lytic development in Salmonella enterica. Virology. 2006;351:360–67.
Hänninen AL, Bamford DH, Bamford JK. Assembly of membrane-containing bacteriophage PRD1 is dependent on GroEL and GroES. Virology. 1997;227:207–10.
Leskinen K, Blasdel BG, Lavigne R, Skurnik M. RNA-sequencing reveals the progression of phage-host interactions between fR1-37 and Yersinia enterocolitica. Viruses. 2016;8:111.
Rossmann FS, Racek T, Wobser D, Puchalka J, Rabener EM, Reiger M, et al. Phage-mediated dispersal of biofilm and distribution of bacterial virulence genes is induced by quorum sensing. PLoS Pathog. 2015;11:e1004653.
Rice SA, Tan CH, Mikkelsen PJ, Kung V, Woo J, Tay M, et al. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 2009;3:271–82.
Davies EV, James CE, Kukavica-Ibrulj I, Levesque RC, Brockhurst MA, Winstanley C. Temperate phages enhance pathogen fitness in chronic lung infection. ISME J. 2016;10:2553–55.
Schuch R, Fischetti VA. The secret life of the anthrax agent Bacillus anthracis: bacteriophage-mediated ecological adaptations. PLoS ONE. 2009;4:e6532.
Schuch R, Fischetti VA. Detailed genomic analysis of the Wbeta and gamma phages infecting Bacillus anthracis: implications for evolution of environmental fitness and antibiotic resistance. J Bacteriol. 2006;188:3037–51.
Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, et al. Cryptic prophages help bacteria cope with adverse environments. Nat Commun. 2010;1:147.
Hosseinidoust Z, Tufenkji N, van de Ven TG. Formation of biofilms under phage predation: considerations concerning a biofilm increase. Biofouling. 2013a;29:457–68.
Abedon ST. Spatial vulnerability: bacterial arrangements, microcolonies, and biofilms as responses to low rather than high phage densities. Viruses. 2012;4:663–87.
Abedon ST. Bacteriophage exploitation of bacterial biofilms: phage preference for less mature targets? FEMS Microbiol Lett. 2016;363:fnv246.
Abedon ST. Phage “delay” towards enhancing bacterial escape from biofilms: a more comprehensive way of viewing resistance to bacteriophages. AIMS Microbiol. 2017;3:186–26.
Gödeke J, Paul K, Lassak J, Thormann KM. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J. 2011;5:613–26.
Binnenkade L, Teichmann L, Thormann KM. Iron triggers λSo prophage induction and release of extracellular DNA in Shewanella oneidensis MR-1 biofilms. Appl Environ Microbiol. 2014;80:5304–16.
Liu X, Li Y, Guo Y, Zeng Z, Li B, Wood TK, et al. Physiological function of rac prophage during biofilm formation and regulation of rac excision in Escherichia coli K-12. Sci Rep. 2015;5:16074.
Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O’Toole GA. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191:210–19.
Tan D, Dahl A, Middelboe M. Vibriophages differentially influence biofilm formation by Vibrio anguillarum strains. Appl Environ Microbiol. 2015;81:4489–97.
Høyland-Kroghsbo NM, Maerkedahl RB, Svenningsen SL. A quorum-sensing-induced bacteriophage defense mechanism. mBio. 2013;4:e00362–12.
Tan D, Svenningsen SL, Middelboe M. Quorum sensing determines the choice of antiphage defense strategy in Vibrio anguillarum. mBio. 2015;6:e00627.
Hoque MM, Naser IB, Bari SM, Zhu J, Mekalanos JJ, Faruque SM. Quorum regulated resistance of Vibrio cholerae against environmental bacteriophages. Sci Rep. 2016;6:37956.
Høyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy-Denomy J, et al. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc Natl Acad Sci USA. 2017;114:131–35.
Gao R, Krysciak D, Petersen K, Utpatel C, Knapp A, Schmeisser C, et al. Genome-wide RNA sequencing analysis of quorum sensing-controlled regulons in the plant-associated Burkholderia glumae PG1 strain. Appl Environ Microbiol. 2015;81:7993–8007.
Patterson AG, Jackson SA, Taylor C, Evans GB, Salmond GP, Przybilski R, et al. Quorum sensing controls adaptive immunity through the regulation of multiple CRISPR-Cas systems. Mol Cell. 2016;64:1102–8.
Winter C, Bouvier T, Weinbauer MG, Thingstad TF. Trade-offs between competition and defense specialists among unicellular planktonic organisms: the “killing the winner” hypothesis revisited. Microbiol Mol Biol Rev. 2010;74:42–57.
Hargreaves KR, Kropinski AM, Clokie MRJ. What does the talking? Quorum sensing signalling genes discovered in a bacteriophage genome. PLoS ONE. 2014;9:e85131.
Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, et al. Communication between viruses guides lysis-lysogeny decisions. Nature. 2017;541:488–93.
Gillis A, Mahillon J. Influence of lysogeny of Tectiviruses GIL01 and GIL16 on Bacillus thuringiensis growth, biofilm formation, and swarming motility. Appl Environ Microbiol. 2014;80:7620–30.
Carrolo M, Frias MJ, Pinto FR, Melo-Cristino J, Ramirez M. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS ONE. 2010;5:e15678.
Acknowledgments
Our work on bacteriophages was funded by grants AGL2015-65673-R (Ministry of Science and Innovation, Spain), EU ANIWHA ERA-NET BLAAT, GRUPIN14-139 (Program of Science, Technology and Innovation 2013-2017 and FEDER EU funds, Principado de Asturias, Spain). L.F. was awarded a “Marie Curie Clarin-Cofund” grant. P.G. and A.R. are members of the bacteriophage network FAGOMAII and the FWO Vlaanderen funded “Phagebiotics” research community (WO.016.14).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Fernández, L., Rodríguez, A. & García, P. Phage or foe: an insight into the impact of viral predation on microbial communities. ISME J 12, 1171–1179 (2018). https://doi.org/10.1038/s41396-018-0049-5
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41396-018-0049-5
This article is cited by
-
Darkness to Discovery: A Comprehensive Mini-Review on Culturable and Non-Culturable Microbial Diversity from Deep Sea
Microbial Ecology (2025)
-
Proteomic analysis reveals phage-driven metabolic shifts and biofilm disruption in methicillin-resistant Staphylococcus aureus (MRSA)
World Journal of Microbiology and Biotechnology (2025)
-
Description of two novel bacteriophages of the class Caudoviricetes that infect Ralstonia solanacearum and Ralstonia pseudosolanacearum
Archives of Virology (2025)
-
The dynamics of the gut microbiota in prediabetes during a four-year follow-up among European patients—an IMI-DIRECT prospective study
Genome Medicine (2025)
-
Phage predation accelerates the spread of plasmid-encoded antibiotic resistance
Nature Communications (2024)