Abstract
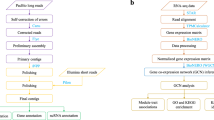
The advent of high-throughput ‘omics approaches coupled with computational analyses to reconstruct individual genomes from metagenomes provides a basis for species-resolved functional studies. Here, a mutual information approach was applied to build a gene association network of a commensal consortium, in which a unicellular cyanobacterium Thermosynechococcus elongatus BP1 supported the heterotrophic growth of Meiothermus ruber strain A. Specifically, we used the context likelihood of relatedness (CLR) algorithm to generate a gene association network from 25 transcriptomic datasets representing distinct growth conditions. The resulting interspecies network revealed a number of linkages between genes in each species. While many of the linkages were supported by the existing knowledge of phototroph-heterotroph interactions and the metabolism of these two species several new interactions were inferred as well. These include linkages between amino acid synthesis and uptake genes, as well as carbohydrate and vitamin metabolism, terpenoid metabolism and cell adhesion genes. Further topological examination and functional analysis of specific gene associations suggested that the interactions are likely to center around the exchange of energetically costly metabolites between T. elongatus and M. ruber. Both the approach and conclusions derived from this work are widely applicable to microbial communities for identification of the interactions between species and characterization of community functioning as a whole.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Di Cagno R, De Angelis M, Calasso M, Gobbetti M. Proteomics of the bacterial cross-talk by quorum sensing. J Proteom. 2011;74:19–34.
Little AE, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62:375–401.
Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33:206–24.
Pande S, Shitut S, Freund L, Westermann M, Bertels F, Colesie C, et al. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun. 2015;6:6238.
Benomar S, Ranava D, Cardenas ML, Trably E, Rafrafi Y, Ducret A, et al. Nutritional stress induces exchange of cell material and energetic coupling between bacterial species. Nat Commun. 2015;6:6283.
Cazelles K, Araujo M, Mouquet M, Gravel D. A theory for species co-occurance in interaction networks. Theor Ecol. 2016;9:39–48.
Harris D. Inferring species interactions from co-occurance data with Markov networks. Cold Spring Harbor Laboratory. 2015.
Madeo D, Comolli LR, Mocenni C. Emergence of microbial networks as response to hostile environments. Front Microbiol. 2014;5:407.
Melnicki MR, Pinchuk GE, Hill EA, Kucek LA, Stolyar SM, Fredrickson JK, et al. Feedback-controlled LED photobioreactor for photophysiological studies of cyanobacteria. Bioresour Technol. 2013;134:127–33.
Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, et al. Initial community evenness favours functionality under selective stress. Nature. 2009;458:623–6.
Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat Biotechnol. 2010;28:245–8.
Biggs MB, Medlock GL, Kolling GL, Papin JA. Metabolic network modeling of microbial communities. Wiley Interdiscip Rev Syst Biol Med. 2015;7:317–34.
Cardona C, Weisenhorn P, Henry C, Gilbert JA. Network-based metabolic analysis and microbial community modeling. Curr Opin Microbiol. 2016;31:124–31.
Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5:93–121.
Borenstein E, Feldman MW. Topological signatures of species interactions in metabolic networks. J Comput Biol. 2009;16:191–200.
Levy R, Borenstein E. Reverse ecology: from systems to environments and back. Adv Exp Med Biol. 2012;751:329–45.
Ishchukov I, Wu Y, Van Puyvelde S, Vanderleyden J, Marchal K. Inferring the relation between transcriptional and posttranscriptional regulation from expression compendia. BMC Microbiol. 2014;14:14.
McClure RS, Overall CC, Mcdermott JE, Hill EA, Markillie LM, McCue LA, et. al. Network analysis of transcriptomics expands regulatory landscapes in Synechococcus sp. PCC 7002. Nucleic Acids Res. 2016;44(18):8810–8825.
Netotea S, Sundell D, Street NR, Hvidsten TR. ComPlEx: conservation and divergence of co-expression networks in A. thaliana, Populus and O. sativa. BMC Genom. 2014;15:106.
Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8.
Van Dam S, Vosa U, Van Der Graaf A, Franke L, De Magalhaes JP. Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform. 2017;p.1–18.
Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinforma. 2005;6:227.
McDermott JE, Oehmen CS, McCue LA, Hill E, Choi DM, Stockel J, et al. A model of cyclic transcriptomic behavior in the cyanobacterium Cyanothece sp. ATCC 51142. Mol Biosyst. 2011;7:2407–18.
Wang J, Wu G, Chen L, Zhang W. Cross-species transcriptional network analysis reveals conservation and variation in response to metal stress in cyanobacteria. BMC Genom. 2013;14:112.
Musungu BM, Bhatnagar D, Brown RL, Payne GA, G OB, Fakhoury AM, et al. A Network Approach of Gene Co-expression in the Zea mays/Aspergillus flavus Pathosystem to Map Host/Pathogen Interaction Pathways. Front Genet. 2016;7:206.
Tierney L, Linde J, Muller S, Brunke S, Molina JC, Hube B, et al. An Interspecies Regulatory Network Inferred from Simultaneous RNA-seq of Candida albicans Invading Innate Immune Cells. Front Microbiol. 2012;3:85.
Bernstein HC, Mcclure R, Thiel V, Sadler N, Kim YM, Chrisler WB, et. al. Indirect interspecies regulation; transcriptional and physiological responses of a cyanobacterium to heterotrophic partnership. mSystems. 2017;2.
Henry CS, Bernstein HC, Weisenhorn P, Taylor RC, Lee JY, Zucker J, et al. Microbial community metabolic modeling: a community data-driven network reconstruction. J Cell Physiol. 2016;231:2339–45.
Iwai M, Katoh H, Katayama M, Ikeuchi M. Improved genetic transformation of the thermophilic cyanobacterium, Thermosynechococcus elongatus BP-1. Plant Cell Physiol. 2004;45:171–5.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Marbach D, Costello JC, Kuffner R, Vega NM, Prill RJ, Camacho DM, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Khuri S, Wuchty S. Essentiality and centrality in protein interaction networks revisited. BMC Bioinforma. 2015;16:109.
Pang K, Sheng H, Ma X. Understanding gene essentiality by finely characterizing hubs in the yeast protein interaction network. Biochem Biophys Res Commun. 2010;401:112–6.
Yang L, Wang J, Wang H, Lv Y, Zuo Y, Jiang W. Characterization of essential genes by topological properties in the perturbation sensitivity network. Biochem Biophys Res Commun. 2014;448:473–9.
McDermott JE, Diamond DL, Corley C, Rasmussen AL, Katze MG, Waters KM. Topological analysis of protein co-abundance networks identifies novel host targets important for HCV infection and pathogenesis. BMC Syst Biol. 2012;6:28.
McDermott JE, Taylor RC, Yoon H, Heffron F. Bottlenecks and hubs in inferred networks are important for virulence in Salmonella typhimurium. J Comput Biol. 2009;16:169–80.
Song HS, McClure RS, Bernstein HC, Overall CC, Hill EA, Beliaev AS. Integrated in silico analyses of regulatory and metabolic networks of Synechococcus sp. PCC 7002 reveal relationships between gene centrality and essentiality. Life. 2015;5:1127–40.
Beliaev AS, Romine MF, Serres M, Bernstein HC, Linggi BE, Markillie LM, et al. Inference of interactions in cyanobacterial-heterotrophic co-cultures via transcriptome sequencing. ISME J. 2014;8:2243–55.
Burgess ML, Barrow KD, Gao C, Heard GM, Glenn D. Carotenoid glycoside esters from the thermophilic bacterium meiothermus ruber. J Nat Prod. 1999;62:859–63.
Hirsch AM. Role of lectins (and rhizobial exopolysaccharides) in legume nodulation. Curr Opin Plant Biol. 1999;2:320–6.
Lerouge I, Vanderleyden J. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol Rev. 2002;26:17–47.
Levy SF, Siegal ML. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol. 2008;6:e264.
Zotenko E, Mestre J, O’leary DP, Przytycka TM. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput Biol. 2008;4:e1000140.
Romine MF, Rodionov DA, Maezato Y, Anderson LN, Nandhikonda P, Rodionova IA, et al. Elucidation of roles for vitamin B12 in regulation of folate, ubiquinone, and methionine metabolism. Proc Natl Acad Sci USA. 2017;114:E1205–14.
Acknowledgements
The authors wish to acknowledge Drs. Margie Romine, William Nelson, and Jim Fredrickson for help with the functional genome annotation and manuscript. PNNL is operated for the DOE by Battelle Memorial Institute under Contract DE-AC05-76RLO 1830. The research was supported by the Genomic Science Program (GSP), Office of Biological and Environmental Research (BER), U.S. Department of Energy (DOE), and is a contribution of the PNNL Foundational Scientific Focus Area (FSFA). A significant portion of the research was performed using the Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by DOE BER and located at PNNL. HCB is grateful for the support of the Linus Pauling Distinguished Postdoctoral Fellowship program at PNNL.
Author contributions
RSM performed the experiments, analyzed the data and wrote the manuscript, CCO analyzed the data and contributed to the manuscript preparation, EAH contributed to the experiments, MC contributed to the experiments, HCB contributed to the experiments and manuscript preparation, JM contributed to the experimental design and manuscript preparation, ASB developed the project concept, contributed to the experimental design, and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
41396_2018_145_MOESM10_ESM.xlsx
Supplementary Table 5: Functional enrichment of T. elongatus genes linked to uncharacterized M. ruber genes SY28_RS04830 and SY28_RS05160
Rights and permissions
About this article
Cite this article
McClure, R.S., Overall, C.C., Hill, E.A. et al. Species-specific transcriptomic network inference of interspecies interactions. ISME J 12, 2011–2023 (2018). https://doi.org/10.1038/s41396-018-0145-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-018-0145-6
This article is cited by
-
Computational systems biology in disease modeling and control, review and perspectives
npj Systems Biology and Applications (2022)
-
TbasCO: trait-based comparative ‘omics identifies ecosystem-level and niche-differentiating adaptations of an engineered microbiome
ISME Communications (2022)
-
Gene co-expression network analysis in zebrafish reveals chemical class specific modules
BMC Genomics (2021)
-
Integrated network modeling approach defines key metabolic responses of soil microbiomes to perturbations
Scientific Reports (2020)
-
Computational Analysis of Interactions of the Oral Microbiota
Current Oral Health Reports (2019)