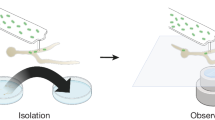
Abstract
Endosymbiosis is a widespread phenomenon in the microbial world and can be based on diverse interactions between endosymbiont and host cell. The vast majority of the known endosymbiotic interactions involve bacteria that have invaded eukaryotic host cells. However, methanogenic archaea have been found to thrive in anaerobic, hydrogenosome-containing protists and it was suggested that this symbiosis is based on the transfer of hydrogen. Here, we used culture-independent genomics approaches to sequence the genomes of two distantly related methanogenic endosymbionts that have been acquired in two independent events by closely related anaerobic ciliate hosts Nyctotherus ovalis and Metopus contortus, respectively. The sequences obtained were then validated as originating from the ciliate endosymbionts by in situ probing experiments. Comparative analyses of these genomes and their closest free-living counterparts reveal that the genomes of both endosymbionts are in an early stage of adaptation towards endosymbiosis as evidenced by the large number of genes undergoing pseudogenization. For instance, the observed loss of genes involved in amino acid biosynthesis in both endosymbiont genomes indicates that the endosymbionts rely on their hosts for obtaining several essential nutrients. Furthermore, the endosymbionts appear to have gained significant amounts of genes of potentially secreted proteins, providing targets for future studies aiming to elucidate possible mechanisms underpinning host-interactions. Altogether, our results provide the first genomic insights into prokaryotic endosymbioses from the archaeal domain of life.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–30.
Ettema TJ. Evolution: mitochondria in the second act. Nature. 2016;531:39–40.
Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–81.
Roger AJ, Muñoz-Gómez SA, Kamikawa R. The origin and diversification of mitochondria. Curr Biol. 2017;27:R1177–92.
Gijzen HJ, Broers C, Barughare M, Stumm CK. Methanogenic bacteria as endosymbionts of the ciliate Nyctotherus ovalis in the cockroach hindgut. Appl Environ Microbiol. 1991;57:1630–4.
Hackstein JH. (Endo)symbiotic methanogenic archaea. Berlin: Springer Science & Business Media; 2010.
van Bruggen JJ, Stumm CK, Zwart KB, Vogels GD. Endosymbiotic methanogenic bacteria of the sapropelic amoeba Mastigella. FEMS Microbiol Lett. 1985;31:187–92.
Embley TM, Finlay B. Systematic and morphological diversity of endosymbiotic methanogens in anaerobic ciliates. Antonie Van Leeuwenhoek. 1993;64:261–71.
Akhmanova A, Voncken F, van Alen T, van Hoek A, Boxma B, Vogels G, et al. A hydrogenosome with a genome. Nature. 1998;396:527–8.
Embley TM, Finlay BJ, Dyal PL, Hirt RP, Wilkinson M, Williams AG. Multiple origins of anaerobic ciliates with hydrogenosomes within the radiation of aerobic ciliates. Proc R Soc Lond B. 1995;262:87–93.
Fenchel T, Finlay B. Production of methane and hydrogen by anaerobic ciliates containing symbiotic methanogens. Arch Microbiol. 1992;157:475–80.
van Bruggen JJ, Zwart KB, van Assema RM, Stumm CK, Vogels GG. Methanobacterium formicicum, an endosymbiont of the anaerobic ciliate Metopus striatus McMurrich. Arch Microbiol. 1984;139:1–7.
Hackstein J, de Graaf R, van Hellemond J, Tielens A. Hydrogenosomes of anaerobic ciliates. In: Tachezy J, editor. Hydrogenosomes and mitosomes: mitochondria of anaerobic eukaryotes. Berlin: Springer-Verlag; 2008. pp. 97–112.
Embley TM, Finlay BJ, Brown S. RNA sequence analysis shows that the symbionts in the ciliate Metopus contortus are polymorphs of a single methanogen species. FEMS Microbiol Lett. 1992;97:57–61.
Finlay B, Fenchel T. Polymorphic bacterial symbionts in the anaerobic ciliated protozoon Metopus. FEMS Microbiol Lett. 1991;79:187–90.
Esteban G, Fenchel T, Finlay B. Diversity of free-living morphospecies in the ciliate genus Metopus. Arch für Protistenkd. 1995;146:137–64.
Finlay B, Fenchel T. Hydrogenosomes in some anaerobic protozoa resemble mitochondria. FEMS Microbiol Lett. 1989;65:311–4.
Esteban G, Guhl BE, Clarke KJ, Embley TM, Finlay BJ. Cyclidium porcatum n. sp.: a free-living anaerobic scuticociliate containing a stable complex of hydrogenosomes, eubacteria and archaeobacteria. Eur J Protistol. 1993;29:262–70.
Goosen NK, Wagener S, Stumm CK. A comparison of two strains of the anaerobic ciliate Trimyema compressum. Arch Microbiol. 1990;153:187–92.
Gutiérrez G, Chistyakova LV, Villalobo E, Kostygov AY, Frolov AO. Identification of Pelomyxa palustris Endosymbionts. Protist. 2017;168:408–24.
Attwood G, Kelly W, Altermann E, Leahy S. Analysis of the Methanobrevibacter ruminantium draft genome: understanding methanogen biology to inhibit their action in the rumen. Aust J Exp Agric. 2008;48:83–8.
Rea S, Bowman JP, Popovski S, Pimm C, Wright A-DG. Methanobrevibacter millerae sp. nov. and Methanobrevibacter olleyae sp. nov., methanogens from the ovine and bovine rumen that can utilize formate for growth. Int J Syst Evolut Microbiol. 2007;57:450–6.
Wright A-DG, Ma X, Obispo NE. Methanobrevibacter phylotypes are the dominant methanogens in sheep from Venezuela. Microb Ecol. 2008;56:390–4.
Miller TL, Wolin M, de Macario EC, Macario A. Isolation of Methanobrevibacter smithii from human feces. Appl Environ Microbiol. 1982;43:227–32.
Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA. 2007;104:10643–8.
Finlay B, Esteban G, Clarke K, Williams A. Some rumen ciliates have endosymbiotic methanogens. FEMS Microbiol Lett. 1994;117:157–61.
Hackstein J, Stumm C. Methane production in terrestrial arthropods. Proc Natl Acad Sci USA. 1994;91:5441–5.
Broers CA, Meijers HH, Symens JC, Stumm CK, Vogels GD. Symbiotic association of Psalteriomonas vulgaris n. sp. with Methanobacterium formicicum. Eur J Protistol. 1993;29:98–105.
Goosen NK, Horemans AM, Hillebrand SJ, Stumm CK, Vogels GD. Cultivation of the sapropelic ciliate Plagiopyla nasuta Stein and isolation of the endosymbiont Methanobacterium formicicum. Arch Microbiol. 1988;150:165–70.
Tokura M, Tajima K, Ushida K. Isolation of Methanobrevibacter sp. as a ciliate-associated ruminal methanogen. J General Appl Microbiol. 1999;45:43–7.
Van Bruggen J, Zwart K, Hermans J, Van Hove E, Stumm CK, Vogels GD. Isolation and characterization of Methanoplanus endosymbiosus sp. nov., an endosymbiont of the marine sapropelic ciliate Metopus contortus Quennerstedt. Arch Microbiol. 1986;144:367–74.
Embley TM, Finlay BJ. The use of small subunit rRNA sequences to unravel the relationships between anaerobic ciliates and their methanogen endosymbionts. Microbiology. 1994;140:225–35.
McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26.
Boscaro V, Felletti M, Vannini C, Ackerman MS, Chain PSG, Malfatti S, et al. Polynucleobacter necessarius, a model for genome reduction in both free-living and symbiotic bacteria. Proc Natl Acad Sci USA. 2013;110:18590–5.
Toft C, Andersson SG. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet. 2010;11:465–75.
Matthews RG. Cobalamin-dependent methyltransferases. Acc Chem Res. 2001;34:681–9.
Khelaifia S, Garibal M, Robert C, Raoult D, Drancourt M. Draft genome sequencing of Methanobrevibacter oralis strain JMR01, isolated from the human intestinal microbiota. Genome Announc. 2014;2:e00073-14.
Ferrari A, Brusa T, Rutili A, Canzi E, Biavati B. Isolation and characterization of Methanobrevibacter oralis sp. nov. Curr Microbiol. 1994;29:7–12.
McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106:15394–9.
Izawa K, Kuwahara H, Kihara K, Yuki M, Lo N, Itoh T, et al. Comparison of intracellular “Ca. Endomicrobium Trichonymphae” genomovars illuminates the requirement and decay of defense systems against foreign DNA. Genome Biol Evol. 2016;8:3099–107.
Yang JC, Madupu R, Durkin AS, Ekborg NA, Pedamallu CS, Hostetler JB, et al. The complete genome of Teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (Shipworms). PLoS ONE. 2009;4:e6085.
Schmitz-Esser S, Tischler P, Arnold R, Montanaro J, Wagner M, Rattei T, et al. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J Bacteriol. 2010;192:1045–57.
Dale C, Plague GR, Wang B, Ochman H, Moran NA. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc Natl Acad Sci USA. 2002;99:12397–402.
Al-Khodor S, Price CT, Kalia A, Kwaik YA. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. 2010;18:132–9.
Jernigan KK, Bordenstein SR. Ankyrin domains across the Tree of Life. PeerJ. 2014;2:e264.
Schmitz-Esser S, Linka N, Collingro A, Beier CL, Neuhaus HE, Wagner M, et al. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J Bacteriol. 2004;186:683–91.
van Hoek AHAM, van Alen TA, Sprakel VSI, Leunissen JAM, Brigge T, Vogels GD, et al. Multiple acquisition of methanogenic archaeal symbionts by anaerobic ciliates. Mol Biol Evol. 2000;17:251–8.
Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93:2873–8.
Fenchel T, Finlay BJ. The biology of free-living anaerobic ciliates. Eur J Protistol. 1991b;26:201–15.
Lewis WH, Sendra KM, Embley TM, Esteban GF. Morphology and phylogeny of a new species of anaerobic ciliate, Trimyema finlayi n. sp., with endosymbiotic methanogens. Front Microbiol. 2018;9:140.
Hoek AHAMV, Sprakel VSI, Alen TAV, Theuvenet APR, Vogels GD, Hackstein JHP. Voltage-dependent reversal of anodic galvanotaxis in Nyctotherus ovalis. J Eukaryot Microbiol. 1999;46:427–33.
Stahl A, Amann R. Development and application of nucleic acid probes. Nucleic acid techniques in bacterial systematics. Chichester: John Wiley & Sons;1991. pp. 205–48.
Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–43.
Daims H, Stoecker K, Wagner M. Fluorescence in situ hybridization for the detection of prokaryotes. Mol Microb Ecol. 2005;213:239.
Ovreås L, Forney L, Daae FL, Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–73.
Jorgensen SL, Hannisdal B, Lanzén A, Baumberger T, Flesland K, Fonseca R, et al. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc Natl Acad Sci USA. 2012;109:E2846–55.
Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
van Hoek AH, van Alen TA, Sprakel VS, Hackstein JH, Vogels GD. Evolution of anaerobic ciliates from the gastrointestinal tract: phylogenetic analysis of the ribosomal repeat from Nyctotherus ovalis and its relatives. Mol Biol Evol. 1998;15:1195–206.
Ellegaard KM, Klasson L, Näslund K, Bourtzis K, Andersson SGE. Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet. 2013;9:e1003381.
St. John, J. SeqPrep. 2011. https://github.com/jstjohn/SeqPrep.
Bolger AM, Lohse M, Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov A, S., et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–5.
Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–9.
Brady A, Salzberg SL. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat Methods. 2009;6:673–6.
Ultsch A, Mörchen F. 2005. ESOM-Maps: tools for clustering, visualization, and classification with Emergent SOM. Marburg: University of Marburg.
Bowers RM, Kyrpides CN, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 2017;35:725–32.
Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119.
Lagesen K, Hallin P, Rødland EA, Stærfeldt H-H, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–8.
Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2011;40:gkr948.
Saw JH, Spang A, Zaremba-Niedzwiedzka K, Juzokaite L, Dodsworth JA, Murugapiran SK, et al. Exploring microbial dark matter to resolve the deep archaeal ancestry of eukaryotes. Philos Trans R Soc B. 2015;370:20140328.
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. Iq-tree: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74.
Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J. ACT: the Artemis comparison tool. Bioinformatics. 2005;21:3422–3.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Gutiérrez G. Draft genome sequence of Methanobacterium formicicum DSM 3637, an archaebacterium isolated from the methane producer amoeba Pelomyxa palustris. J Bacteriol. 2012;194:6967–8.
Fenchel T, Finlay BJ. Ecology and evolution in anoxic worlds. Oxford: Oxford University Press; 1995.
van Bruggen JJ, Stumm CK, Vogels GD. Symbiosis of methanogenic bacteria and sapropelic protozoa. Arch Microbiol. 1983;136:89–95.
Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–9.
Bagos P, Tsirigos K, Plessas S, Liakopoulos T, Hamodrakas S. Prediction of signal peptides in archaea. Protein Eng Des Sel. 2008;22:27–35.
Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80.
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3.
Csűös M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26:1910–2.
Acknowledgements
We thank Prof. Genoveva Esteban for help in isolating and identifying cultures of M. contortus, and we are grateful to Dr. Nicole Poulton for the initial evaluation of N. ovalis lysates. We would also like to thank Roel van Eijk and Lina Juzokaite for their help with sequencing library construction and Daniel Lundblad for his help in establishing cockroach cultivation. All sequencing was performed by the National Genomics Infrastructure sequencing platforms at the Science for Life Laboratory at Uppsala University, a national infrastructure supported by the Swedish Research Council (VR-RFI) and the Knut and Alice Wallenberg Foundation. We thank the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) at Uppsala University and the Swedish National Infrastructure for Computing (SNIC) at the PDC Center for High-Performance Computing for providing computational resources. This work was supported by grants of the Swedish Research Council (VR grant 621-2009-4813), the European Research Council (ERC Starting grant 310039-PUZZLE_CELL) and the Swedish Foundation for Strategic Research (SSF-FFL5) to TJGE and to TME (ERC Advanced grant 20100317-EUKORIGINMIT).
Author contributions
TJGE conceived and supervised the study. AEL isolated the endosymbionts and performed genomic sequence assemblies and analysis. AEL performed comparative genomics and phylogenomics analyses with occasional support from LG. WHL and TME isolated and cultured M. contortus samples and performed the fluorescent in situ hybridization experiments, as well as microscopic imaging. AS performed the metabolic analysis. AEL and TJGE wrote the manuscript, which was edited and approved by all authors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lind, A.E., Lewis, W.H., Spang, A. et al. Genomes of two archaeal endosymbionts show convergent adaptations to an intracellular lifestyle. ISME J 12, 2655–2667 (2018). https://doi.org/10.1038/s41396-018-0207-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-018-0207-9
This article is cited by
-
Epibiotic Microbiomes Dominated by Pseudoalteromonas Were Associated with the Unicellular Ciliate Paraspathidium apofuscum from Marine Sediments
Journal of Ocean University of China (2026)
-
Distinct evolutionary origins and mixed-mode transmissions of methanogenic endosymbionts are revealed in anaerobic ciliated protists
Marine Life Science & Technology (2025)
-
Genetic potential for aerobic respiration and denitrification in globally distributed respiratory endosymbionts
Nature Communications (2024)
-
The parasitic lifestyle of an archaeal symbiont
Nature Communications (2024)
-
Symbionts of Ciliates and Ciliates as Symbionts
Indian Journal of Microbiology (2024)