Abstract
Ecological factors contributing to depth-related diversification of marine Thaumarchaeota populations remain largely unresolved. To investigate the role of potential microbial associations in shaping thaumarchaeal ecotype diversification, we examined co-occurrence relationships in a community composition dataset (16S rRNA V4-V5 region) collected as part of a 2-year time series in coastal Monterey Bay. Ecotype groups previously defined based on functional gene diversity—water column A (WCA), water column B (WCB) and Nitrosopumilus-like clusters—were recovered in the thaumarchaeal 16S rRNA gene phylogeny. Networks systematically reflected depth-related patterns in the abundances of ecotype populations, suggesting thaumarchaeal ecotypes as keystone members of the microbial community below the euphotic zone. Differential environmental controls on the ecotype populations were further evident in subnetwork modules showing preferential co-occurrence of OTUs belonging to the same ecotype cluster. Correlated abundances of Thaumarchaeota and heterotrophic bacteria (e.g., Bacteroidetes, Marinimicrobia and Gammaproteobacteria) indicated potential reciprocal interactions via dissolved organic matter transformations. Notably, the networks recovered ecotype-specific associations between thaumarchaeal and Nitrospina OTUs. Even at depths where WCB-like Thaumarchaeota dominated, Nitrospina OTUs were found to preferentially co-occur with WCA-like and Nitrosopumilus-like thaumarchaeal OTUs, highlighting the need to investigate the ecological implications of the composition of nitrifier assemblages in marine waters.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
Sequence data have been deposited in the NCBI Sequence Read Archive (SRA) database under BioProject PRJNA488312 (SRA accession: SRP159037).
References
Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–10.
Teira E, van Aken H, Veth C, Herndl GJ. Archaeal uptake of enantiomeric amino acids in the meso‐ and bathypelagic waters of the North Atlantic. Limnol Oceanogr. 2006;51:60–69.
Könneke M, Bernhard AE, la Torre de JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–6.
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. PNAS. 2008;105:2134–9.
Santoro AE, Casciotti KL. Enrichment and characterization of ammonia-oxidizing archaea from the open ocean: phylogeny, physiology and stable isotope fractionation. ISME J. 2011;5:1796–808.
Stieglmeier M, Klingl A, Alves RJE, Rittmann SKMR, Melcher M, Leisch N, et al. Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Micro. 2014;64:2738–52.
Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH, et al. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. PNAS. 2014;111:12504–9.
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. PNAS. 2005;102:14683–8.
Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Micro. 2007;9:1162–75.
Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:453–63.
Yakimov MM, Cono VL, Denaro R. A first insight into the occurrence and expression of functional amoA and accA genes of autotrophic and ammonia-oxidizing bathypelagic Crenarchaeota of Tyrrhenian Sea. Deep Sea Res Part II. 2009;56:748–54.
Santoro AE, Casciotti KL, Francis CA. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol. 2010;12:1989–2006.
Tolar BB, King GM, Hollibaugh JT. An analysis of Thaumarchaeota populations from the Northern Gulf of Mexico. Front Microbiol. 2013;4:72. https://doi.org/10.3389/fmicb.2013.00072.
Tolar BB, Ross MJ, Wallsgrove NJ, Liu Q, Aluwihare LI, Popp BN, et al. Contribution of ammonia oxidation to chemoautotrophy in Antarctic coastal waters. ISME J. 2016;10:2605–19.
Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y-I, Sugahara J, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. PNAS. 2006;103:18296–301.
Church MJ, Wai B, Karl DM, DeLong EF. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol. 2010;12:679–88.
Hu A, Jiao N, Zhang CL. Community structure and function of planktonic Crenarchaeota: changes with depth in the South China sea. Microb Ecol. 2011;62:549–63.
Mosier AC, Francis CA. Determining the distribution of marine and coastal ammonia-oxidizing archaea and bacteria using a quantitative approach. In: Martin G. Klotz (ed.) Research on Nitrification and Related Processes, Part A, vol. 486. Elsevier: Methods in Enzymology; 2011, pp. 205–21.
Smith JM, Casciotti KL, Chavez FP, Francis CA. Differential contributions of archaeal ammonia oxidizer ecotypes to nitrification in coastal surface waters. ISME J. 2014;8:1704–14.
Smith JM, Damashek J, Chavez FP, Francis CA. Factors influencing nitrification rates and the abundance and transcriptional activity of ammonia‐oxidizing microorganisms in the dark northeast Pacific Ocean. Limnol Oceanogr. 2016;61:596–609.
Luo H, Sun Y, Hollibaugh JT, Moran MA. Low genome content diversity of marine planktonic Thaumarchaeota. Environ Microbiol Rep. 2016;8:501–7.
Santoro AE, Saito MA, Goepfert TJ, Lamborg CH, Dupont CL, DiTullio GR. Thaumarchaeal ecotype distributions across the equatorial Pacific Ocean and their potential roles in nitrification and sinking flux attenuation. Limnol Oceanogr. 2017;62:1984–2003.
Müller O, Wilson B, Paulsen ML, Rumińska A, Armo HR, Bratbak G, et al. Spatiotemporal dynamics of ammonia-oxidizing Thaumarchaeota in distinct Arctic water masses. Front Microbiol. 2018;9:24.
Luo H, Tolar BB, Swan BK, Zhang CL, Stepanauskas R, Moran MA, et al. Single-cell genomics shedding light on marine Thaumarchaeota diversification. ISME J. 2014;8:732–6.
Merbt SN, Stahl DA, Casamayor EO, Martí E, Nicol GW, Prosser JI. Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett. 2012;327:41–46.
Tolar BB, Powers LC, Miller WL, Wallsgrove NJ, Popp BN, Hollibaugh JT. Ammonia oxidation in the ocean can be inhibited by nanomolar concentrations of hydrogen peroxide. Front Mar Sci. 2016;3:499.
Sintes E, Bergauer K, De Corte D, Yokokawa T, Herndl GJ. Archaeal amoA gene diversity points to distinct biogeography of ammonia-oxidizing Crenarchaeota in the ocean. Enviro Microbiol. 2013;15:1647–58.
Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comp Biol. 2012;8:e1002606.
Layeghifard M, Hwang DM, Guttman DS. Disentangling interactions in the microbiome: a network perspective. Trends Microbiol. 2017;25:217–28.
He S, Guo L, Niu M, Miao F, Jiao S, Hu T, et al. Ecological diversity and co-occurrence patterns of bacterial community through soil profile in response to long-term switchgrass cultivation. Sci Rep. 2017;7:3608.
Fuhrman JA, Steele JA. Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquat Microb Ecol. 2008;53:69–81.
Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 2011;5:1414–25.
Eiler A, Heinrich F, Bertilsson S. Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J. 2012;6:330–42.
Chow C-ET, Sachdeva R, Cram JA, Steele JA, Needham DM, Patel A, et al. Temporal variability and coherence of euphotic zone bacterial communities over a decade in the Southern California Bight. ISME J. 2013;7:2259–73.
Vick-Majors TJ, Priscu JC, Amaral-Zettler LA. Modular community structure suggests metabolic plasticity during the transition to polar night in ice-covered Antarctic lakes. ISME J. 2013;8:778–89.
Cram JA, Xia LC, Needham DM, Sachdeva R, Sun F, Fuhrman JA. Cross-depth analysis of marine bacterial networks suggests downward propagation of temporal changes. ISME J. 2015;9:2573–86.
Baldassano SN, Bassett DS. Topological distortion and reorganized modular structure of gut microbial co-occurrence networks in inflammatory bowel disease. Sci Rep. 2016;6:26087.
Tapio I, Fischer D, Blasco L, Tapio M, Wallace RJ, Bayat AR, et al. Taxon abundance, diversity, co-occurrence and network analysis of the ruminal microbiota in response to dietary changes in dairy cows. Smidt H, editor. PLoS ONE. 2017;12:e0180260.
Parada AE, Fuhrman JA. Marine archaeal dynamics and interactions with the microbial community over 5 years from surface to seafloor. ISME J. 2017;11:2510–25.
Daebeler A, Bodelier PL, Yan Z, Hefting MM, Jia Z, Laanbroek HJ. Interactions between Thaumarchaea, Nitrospira and methanotrophs modulate autotrophic nitrification in volcanic grassland soil. ISME J. 2014;8:2397–410.
Xiao Y, Angulo MT, Friedman J, Waldor MK, Weiss ST, Liu Y-Y. Mapping the ecological networks of microbial communities. Nat Comm. 2017;8:2042.
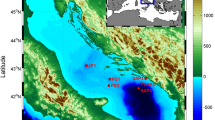
Timothy Pennington J, Chavez FP. Seasonal fluctuations of temperature, salinity, nitrate, chlorophyll and primary production at station H3/M1 over 1989-1996 in Monterey Bay, California. Deep Sea Res Part 2. 2000;47:947–73.
Smith JM, Chavez FP, Francis CA. Ammonium uptake by phytoplankton regulates nitrification in the sunlit ocean. PLoS ONE. 2014;9:e108173.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–6. https://doi.org/10.1093/nar/gks1219
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200.
R CT. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017.
McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217.
Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos P et al. (2008), vegan: Community Ecology Package. R package version 2.5-2.
Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comp Biol. 2015;11:e1004226.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U, et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311:496–503.
Brown MV, Philip GK, Bunge JA, Smith MC, Bissett A, Lauro FM, et al. Microbial community structure in the North Pacific ocean. ISME J. 2009;3:1374–86.
Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3:1148–63.
Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Ben T, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308.
Ghiglione J-F, Galand PE, Pommier T, Pedrós-Alió C, Maas EW, Bakker K, et al. Pole-to-pole biogeography of surface and deep marine bacterial communities. PNAS. 2012;109:17633–88.
Hatosy SM, Martiny JBH, Sachdeva R, Steele J, Fuhrman JA, Martiny AC. Beta diversity of marine bacteria depends on temporal scale. Ecology. 2013;94:1898–904.
Walsh EA, Kirkpatrick JB, Rutherford SD, Smith DC, Sogin M, D’Hondt S. Bacterial diversity and community composition from seasurface to subseafloor. ISME J. 2015;10:978–89.
Yaveroğlu ÖN, Malod-Dognin N, Davis D, Levnajic Z, Janjic V, Karapandza R, et al. Revealing the hidden language of complex networks. Sci Rep. 2014;4:04547.
Peura S, Bertilsson S, Jones RI, Eiler A. Resistant microbial cooccurrence patterns inferred by network topology. Appl Environ Microbiol. 2015;81:2090–97.
Williams RJ, Howe A, Hofmockel KS. Demonstrating microbial co-occurrence pattern analyses within and between ecosystems. Front Microbiol. 2014;5:358.
Fillol M, Auguet J-C, Casamayor EO, Borrego CM. Insights in the ecology and evolutionary history of the Miscellaneous Crenarchaeotic Group lineage. ISME J. 2015;10:665–77.
Banerjee S, Kirkby CA, Schmutter D, Bissett A, Kirkegaard JA, Richardson AE. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol Biochem. 2016;97:188–98.
van der Heijden MGA, Hartmann M. Networking in the plant microbiome. PLoS Biol. 2016;14:e1002378.
Hawley AK, Nobu MK, Wright JJ, Durno WE, Morgan-Lang C, Sage B, et al. Diverse marinimicrobia bacteria may mediate coupled biogeochemical cycles along eco-thermodynamic gradients. Nat Comm. 2017;8:1507.
Santoro AE, Buchwald C, McIlvin MR, Casciotti KL. Isotopic signature of N2O produced by marine ammonia-oxidizing archaea. Science. 2011;333:1282–5.
Martens-Habbena W, Qin W, Horak REA, Urakawa H, Schauer AJ, Moffett JW, et al. The production of nitric oxide by marine ammonia‐oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ Microbiol. 2015;17:2261–74.
Kozlowski JA, Kits KD, Stein LY. Comparison of nitrogen oxide metabolism among diverse ammonia-oxidizing bacteria. Front Microbiol. 2016;7:1703.
Fernández-Gómez B, Richter M, Schüler M, Pinhassi J, Acinas SG, González JM, et al. Ecology of marine bacteroidetes: a comparative genomics approach. ISME J. 2013;7:1026–37.
Bergauer K, Fernandez-Guerra A, Garcia JAL, Sprenger RR, Stepanauskas R, Pachiadaki MG, et al. Organic matter processing by microbial communities throughout the Atlantic water column as revealed by metaproteomics. PNAS. 2017;57:1708779115.
Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–41.
Fuhrman JA. Microbial community structure and its functional implications. Nature. 2009;459:193–9.
Pachiadaki MG, Sintes E, Bergauer K, Brown JM, Record NR, Swan BK, et al. Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science. 2017;358:1046–51.
Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–51.
Ngugi DK, Blom J, Stepanauskas R, Stingl U. Diversification and niche adaptations of Nitrospina-like bacteria in the polyextreme interfaces of Red Sea brines. ISME J. 2016;10:1383–99.
Acknowledgments
We thank Tim Pennington, Marguerite Blum, and other members of the Biological Ocean Group at MBARI for intellectual and logistical support. We also thank the captain & crew of the RV Western Flyer and RV Rachel Carson (MBARI) for support in sample acquisition efforts at sea. Jessica Lee, Emily Cardarelli and Madison Jackson assisted with sample collection; additional assistance with sample processing was done by Tynan Challenor. Sequencing support came from a DOE JGI CSP project to CAF. JGI project manager Tijana Glavina del Rio facilitated sample submission and data acquisition. Field research was supported (in part) by the grant OCE-1357024 from NSF Biological Oceanography (to CAF), an ARCS Foundation Graduate Research Fellowship and an MBARI Postdoctoral Fellowship (to JMS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Reji, L., Tolar, B.B., Smith, J.M. et al. Differential co-occurrence relationships shaping ecotype diversification within Thaumarchaeota populations in the coastal ocean water column. ISME J 13, 1144–1158 (2019). https://doi.org/10.1038/s41396-018-0311-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-018-0311-x
This article is cited by
-
Abundant and metabolically flexible bacterial lineages underlie a vast potential for rubisco-mediated carbon fixation in the dark ocean
Genome Biology (2025)
-
Plant carbon allocation, soil carbon and nutrient condition, and microbial community jointly regulate microbial biomass carbon accumulation
Plant and Soil (2025)
-
Thaumarchaeota from deep-sea methane seeps provide novel insights into their evolutionary history and ecological implications
Microbiome (2024)
-
Biogeographic shifts in the microbial co-occurrence network features of three domains across complex environmental gradients in subtropical coastal waters
Ecological Processes (2024)
-
Elevated hydrostatic pressure enhances the potential for microbially mediated carbon sequestration at the sediment–water interface in a deep-water reservoir by modulating functional genes and metabolic pathways
Carbon Research (2024)