Abstract
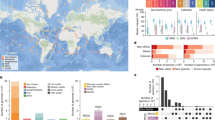
Modeling crude-oil biodegradation in sediments remains a challenge due in part to the lack of appropriate model organisms. Here we report the metagenome-guided isolation of a novel organism that represents a phylogenetically narrow (>97% 16S rRNA gene identity) group of previously uncharacterized, crude-oil degraders. Analysis of available sequence data showed that these organisms are highly abundant in oiled sediments of coastal marine ecosystems across the world, often comprising ~30% of the total community, and virtually absent in pristine sediments or seawater. The isolate genome encodes functional nitrogen fixation and hydrocarbon degradation genes together with putative genes for biosurfactant production that apparently facilitate growth in the typically nitrogen-limited, oiled environment. Comparisons to available genomes revealed that this isolate represents a novel genus within the Gammaproteobacteria, for which we propose the provisional name “Candidatus Macondimonas diazotrophica” gen. nov., sp. nov. “Ca. M. diazotrophica” appears to play a key ecological role in the response to oil spills around the globe and could be a promising model organism for studying ecophysiological responses to oil spills.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
The data reported in this paper are publicly available through the Gulf of Mexico Research Initiative Information & Data Cooperative (GRIIDC), under the accession numbers R5.x278.000:0014 and R5.x278.000:0002 (NCBI accession MH795143). The metagenome sequences as well as the assembled genome sequences are also available at http://enve-omics.ce.gatech.edu/data/.
References
Atlas RM, Hazen TC. Oil biodegradation and bioremediation: a tale of the two worst spills in U.S. history. Environ Sci Technol. 2011;45:6709–15.
Bagby SC, Reddy CM, Aeppli C, Fisher GB, Valentine DL. Persistence and biodegradation of oil at the ocean floor following Deepwater Horizon. Proc Natl Acad Sci USA. 2017;114:E9–e18.
King GM, Kostka JE, Hazen TC, Sobecky PA. Microbial Responses to the Deepwater Horizon Oil Spill: From Coastal Wetlands to the Deep Sea. Annu Rev Mar Sci. 2015;7:377–401.
Mason OU, Hazen TC, Borglin S, Chain PSG, Dubinsky EA, Fortney JL, et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 2012;6:1715.
Handley KM, Piceno YM, Hu P, Tom LM, Mason OU, Andersen GL, et al. Metabolic and spatio-taxonomic response of uncultivated seafloor bacteria following the Deepwater Horizon oil spill. ISME J. 2017;11:2569.
Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204.
Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, et al. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico Beach Sands Impacted by the Deepwater Horizon Oil Spill. Appl Environ Microbiol. 2011;77:7962–74.
Michel J, Owens EH, Zengel S, Graham A, Nixon Z, Allard T, et al. Extent and degree of shoreline oiling: Deepwater Horizon Oil Spill, Gulf of Mexico, USA. PLoS ONE. 2013;8:e65087.
Mendelssohn IA, Andersen GL, Baltz DM, Caffey RH, Carman KR, Fleeger JW, et al. Oil impacts on coastal wetlands: implications for the Mississippi River Delta Ecosystem after the Deepwater Horizon Oil Spill. BioScience. 2012;62:562–74.
Rodriguez-R LM, Overholt WA, Hagan C, Huettel M, Kostka JE, Konstantinidis KT. Microbial community successional patterns in beach sands impacted by the Deepwater Horizon oil spill. ISME J. 2015;9:1928–40.
Huettel M, Overholt WA, Kostka JE, Hagan C, Kaba J, Wells WB et al. Degradation of Deepwater Horizon oil buried in a Florida beach influenced by tidal pumping. Mar Poll Bull. 2018;126:488–500.
Pritchard PH, Mueller JG, Rogers JC, Kremer FV, Glaser JA. Oil spill bioremediation: experiences, lessons and results from the Exxon Valdez oil spill in Alaska. Biodegradation. 1992;3:315–35.
Head IM, Jones DM, Roling WFM. Marine microorganisms make a meal of oil. Nat Rev Micro. 2006;4:173–82.
Huettel M, Overholt WA, Kostka JE, Hagan C, Kaba J, Wells WB, et al. Degradation of Deepwater Horizon oil buried in a Florida beach influenced by tidal pumping. Mar Poll Bull. 2018;126:488–500.
Caro-Quintero A, Konstantinidis KT. Bacterial species may exist, metagenomics reveal. Environ Microbiol. 2012;14:347–55.
Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, et al. Recovery of nearly 8000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–42.
Konstantinidis KT, Rosselló-Móra R, Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399.
Rodriguez-R LM, Gunturu S, Harvey WT, Rosselló-Mora R, Tiedje JM, Cole JR, et al. The Microbial Genomes Atlas (MiGA) webserver: taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018;46:W282–W288.
Slobodkina GB, Baslerov RV, Novikov AA, Viryasov MB, Bonch-Osmolovskaya EA, Slobodkin AI. Inmirania thermothiophila gen. nov., sp. nov., a thermophilic, facultatively autotrophic, sulfur-oxidizing gammaproteobacterium isolated from a shallow-sea hydrothermal vent. Int J Syst Evolut Microbiol. 2016;66:701–6.
Oren A. The Family Ectothiorhodospiraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes: Gammaproteobacteria. Berlin, Heidelberg: Springer Science+Business Media; 2014. p. 199–222.
Watson SW. Taxonomic considerations of the family Nitrobacteraceae Buchanan. Int J Syst Evolut Microbiol. 1971;21:254–70.
Jiang Y, Sorokin DY, Kleerebezem R, Muyzer G, van Loosdrecht M. Plasticicumulans acidivorans gen. nov., sp. nov., a polyhydroxyalkanoate-accumulating gammaproteobacterium from a sequencing-batch bioreactor. Int J Syst Evolut Microbiol. 2011;61:2314–9.
Rodriguez-R LM, Overholt WA, Hagan C, Huettel M, Kostka JE, Konstantinidis KT. Microbial community successional patterns in beach sands impacted by the Deepwater Horizon oil spill. ISME J. 2015;9:1928–1940.
Acknowledgements
The authors would like to thank Dr. Aharon Oren for his valuable input on the naming of the new type species. This research was made possible by a grant from The Gulf of Mexico Research Initiative (Grant No 321611-00; RFP V). Data are publicly available through the Gulf of Mexico Research Initiative Information & Data Cooperative (GRIIDC) at https://data.gulfresearchinitiative.org (UDI: R5.x278.000:0014, UDI: R5.x278.000:0002). RR-M acknowledges the financial support for his sabbatical stay at Georgia Tech from the Spanish Ministry of Sciences, Innovation and Universities (Grant No. PRX18/00048).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Karthikeyan, S., Rodriguez-R, L.M., Heritier-Robbins, P. et al. “Candidatus Macondimonas diazotrophica”, a novel gammaproteobacterial genus dominating crude-oil-contaminated coastal sediments. ISME J 13, 2129–2134 (2019). https://doi.org/10.1038/s41396-019-0400-5
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41396-019-0400-5
This article is cited by
-
Finding microbial composition and biological processes as predictive signature to access the ongoing status of mangrove preservation
International Microbiology (2024)
-
The marine nitrogen cycle: new developments and global change
Nature Reviews Microbiology (2022)
-
Universal activity-based labeling method for ammonia- and alkane-oxidizing bacteria
The ISME Journal (2022)
-
Phylogenomic analysis of a metagenome-assembled genome indicates a new taxon of an anoxygenic phototroph bacterium in the family Chromatiaceae and the proposal of “Candidatus Thioaporhodococcus” gen. nov
Archives of Microbiology (2022)
-
Beach sand oil spills select for generalist microbial populations
The ISME Journal (2021)