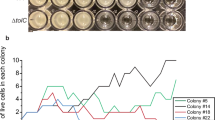
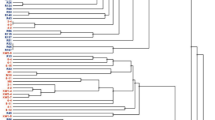
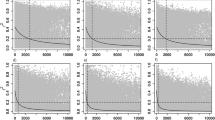
Abstract
High rates of homologous recombination (HR) in the bacterial plant pathogen Xylella fastidiosa have been previously detected. This study aimed to determine the extent and explore the ecological significance of HR in the genomes of recombinants experimentally generated by natural transformation and wild-type isolates. Both sets of strains displayed widespread HR and similar average size of recombined fragments consisting of random events (2–10 kb) of inter- and intrasubspecific recombination. A significantly higher proportion and greater lengths (>10 kb, maximum 31.5 kb) of recombined fragments were observed in subsp. morus and in strains isolated in Europe from intercepted coffee plants shipped from the Americas. Such highly recombinant strains pose a serious risk of emergence of novel variants, as genetically distinct and formerly geographically isolated genotypes are brought in close proximity by global trade. Recently recombined regions in wild-type strains included genes involved in regulation and signaling, host colonization, nutrient acquisition, and host evasion, all fundamental traits for X. fastidiosa ecology. Identification of four recombinant loci shared between wild-type and experimentally generated recombinants suggests potential hotspots of recombination in this naturally competent pathogen. These findings provide insights into evolutionary forces possibly affecting the adaptive potential to colonize the host environments of X. fastidiosa.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–21.
Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001;2(1):376–81.
Michod RE, Bernstein H, Nedelcu AM. Adaptive value of sex in microbial pathogens. Infect, Genet Evol: J Mol Epidemiol Evolut Genet Infect Dis. 2008;8:267–85.
Guttman DS. Recombination and clonality in natural populations of Escherichia coli. Trends Ecol Evol. 1997;12:16–22.
Feil EJ, Spratt BG. Recombination and the population structures of bacterial pathogens. Annu Rev Microbiol. 2001;55:561–90.
Nelson K, Selander RK. Intergeneric transfer and recombination of the 6-phosphogluconate dehydrogenase gene (gnd) in enteric bacteria. Proc Natl Acad Sci USA. 1994;91:10227–31.
Yan S, Liu H, Mohr TJ, Jenrette J, Chiodini R, Zaccardelli M, et al. Role of recombination in the evolution of the model plant pathogen Pseudomonas syringae pv. tomato DC3000, a ver, atypical tomato strain. Appl Environ Microbiol. 2008;74:3171–81.
Baltrus DA, Guillemin K, Phillips PC, Travisano M. Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution. 2008;62:39–49.
Feil EJ, Holmes EC, Bessen DE, Chan MS, Day NP, Enright MC, et al. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci USA. 2001;98:182–7.
Spratt BG, Bowler LD, Zhang QY, Zhou J, Smith JM. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J Mol Evol. 1992;34:115–25.
Maiden MC. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–88.
Freel KC, Millán-Aguiñaga N, Jensen PR. Multilocus sequence typing reveals evidence of homologous recombination linked to antibiotic resistance in the genus Salinispora. Appl Environ Microbiol. 2013;79:5997–6005.
Kong Y, Ma JH, Warren K, Tsang RS, Low DE, Jamieson FB, et al. Homologous recombination drives both sequence diversity and gene content variation in Neisseria meningitidis. Genome Biol Evol. 2013;5:1611–27.
Timilsina S, Jibrin MO, Potnis N, Minsavage GV, Kebede M, Schwartz A, et al. Multilocus sequence analysis of xanthomonads causing bacterial spot of tomato and pepper plants reveals strains generated by recombination among species and recent global spread of Xanthomonas gardneri. Appl Environ Microbiol. 2015;81:1520–9.
Nunney L, Hopkins DL, Morano LD, Russell SE, Stouthamer R. Intersubspecific recombination in Xylella fastidiosa strains native to the United States: infection of novel hosts associated with an unsuccessful invasion. Appl Environ Microbiol. 2014;80:1159–69.
Parker JK, Havird JC, De La, Fuente L. Differentiation of Xylella fastidiosa strains via multilocus sequence analysis of environmentally mediated genes (MLSA-E). Appl Environ Microbiol. 2012;78:1385–96.
Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15
Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, et al. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 2012;40:e6
Yahara K, Didelot X, Ansari MA, Sheppard SK, Falush D. Efficient inference of recombination hot regions in bacterial genomes. Mo Biol Evol. 2014;31:1593–605.
Almeida RP, Nunney L. How do plant diseases caused by Xylella fastidiosa emerge?. Plant Dis. 2015;99:1457–67.
Saponari M, Boscia D, Nigro F, Martelli GP. Identification of DNA sequences related to Xylella fastidiosa in Oleander, Almond and Olive trees exhibiting leaf scorch symptoms in Apulia (Southern Italy). J Plant Pathol. 2013;95:668
Strona G, Carstens CJ, Beck PS. Network analysis reveals why Xylella fastidiosa will persist in Europe. Sci Rep 2017;7:71
Su CC, Deng WL, Jan FJ, Chang CJ, Huang H, Shih HT, et al. Xylella taiwanensis sp. nov., causing pear leaf scorch disease. Int J Syst Evolut Microbiol. 2016;66:4766–71.
Hopkins DL, Purcell AH. Xylella fastidiosa: cause of Pierce’s disease of grapevine and other emergent diseases. Plant Dis. 2002;86:1056–66.
Marcelletti S, Scortichini M. Genome-wide comparison and taxonomic relatedness of multiple Xylella fastidiosa strains reveal the occurrence of three subspecies and a new Xylella species. Arch Microbiol. 2016;198:803–12.
Schaad NW, Postnikova E, Lacy G, Fatmi M, Chang CJ. Xylella fastidiosa subspecies: X. fastidiosa subsp piercei, subsp. nov., X. fastidiosa subsp. multiplex subsp. nov., and X. fastidiosa subsp. pauca subsp. nov. Syst Appl Microbiol. 2004;27:763
Schaad NW, Postnikova E, Lacy G, Fatmi MB, Chang CJ. Xylella fastidiosa subspecies: X-fastidiosa subsp piercei, subsp nov., X-fastidiosa subsp multiplex subsp. nov., and X-fastidiosa subsp pauca subsp nov. Syst Appl Microbiol. 2004;27:290–300.
Nunney L, Schuenzel EL, Scally M, Bromley RE, Stouthamer R. Large-scale intersubspecific recombination in the plant-pathogenic bacterium Xylella fastidiosa is associated with the host shift to mulberry. Appl Environ Microbiol. 2014;80:3025–33.
Scally M, Schuenzel EL, Stouthamer R, Nunney L. Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Appl Environ Microbiol. 2005;71:8491–9.
EFSA. Update of the Xylella spp. host plant database. EFSA Journal. 2018;16:5408.
Nunney L, Yuan X, Bromley RE, Stouthamer R. Detecting genetic introgression: high levels of intersubspecific recombination found in Xylella fastidiosa in Brazil. Appl Environ Microbiol. 2012;78:4702–14.
Almeida RP, Nascimento FE, Chau J, Prado SS, Tsai CW, Lopes SA, et al. Genetic structure and biology of Xylella fastidiosa strains causing disease in citrus and coffee in Brazil. Appl Environ Microbiol. 2008;74:3690–701.
Coletta-Filho H, Francisco C, Lopes J, Muller C, Almeida R. Homologous recombination and Xylella fastidiosa host-pathogen associations in South America. Phytopathology. 2017;107:305–312.
Nunney L, Yuan X, Bromley R, Hartung J, Montero-Astua M, Moreira L, et al. Population genomic analysis of a bacterial plant pathogen: novel insight into the origin of Pierce’s disease of grapevine in the U.S. PLoS One. 2010;5:e15488
Kung SH, Almeida RP. Natural competence and recombination in the plant pathogen Xylella fastidiosa. Appl Environ Microbiol. 2011;77:5278–84.
Kandel PP, Lopez SM, Almeida RP, De La Fuente L. Natural competence of Xylella fastidiosa occurs at a high frequency inside microfluidic chambers mimicking the bacterium’s natural habitats. Appl Environ Microbiol. 2016;82:5269–77.
Kandel PP, Almeida RPP, Cobine PA, De La Fuente L. Natural competence rates are variable among Xylella fastidiosa strains and homologous recombination occurs in vitro between subspecies fastidiosa and multiplex. Mol Plant-Microbe Interact. 2017;30:589–600.
Kandel PP, Chen HY, De La Fuente L. A short protocol for gene knockout and complementation in Xylella fastidiosa shows that one of the type IV pilin paralogs (PD1926) is needed for twitching while another (PD1924) affects pilus number and location. Appl Environ Microbiol. 2018;84:e01167–18.
Oliver JE, Sefick SA, Parker JK, Arnold T, Cobine PA, De La Fuente L. Ionome changes in Xylella fastidiosa-infected Nicotiana tabacum correlate with virulence and discriminate between subspecies of bacterial isolates. Mol Plant-Microbe Interact. 2014;27:1048–58.
Bergsma-Vlami M, van de Bilt JLJ, Tjou-Tam-Sin NNA, Helderman CM, Gorkink-Smits P, Landman NM, et al. Assessment of the genetic diversity of Xylella fastidiosa in imported ornamental Coffea arabica plants. Plant Pathol. 2017;66:1065–74.
Davis MJ, French WJ, Schaad NW. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr Microbiol. 1981;6:309–14.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147.
Cheng L, Connor TR, Siren J, Aanensen DM, Corander J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol. 2013;30:1224–8.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics . 2014;30:1312–3.
Mostowy R, Croucher NJ, Andam CP, Corander J, Hanage WP, Marttinen P. Efficient inference of recent and ancestral recombination within bacterial populations. Mol Biol Evol. 2017;34:1167–82.
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3.
Bubendorfer S, Krebes J, Yang I, Hage E, Schulz TF, Bahlawane C, et al. Genome-wide analysis of chromosomal import patterns after natural transformation of Helicobacter pylori. Nat Commun. 2016;7:11995-
Zhang S, Chakrabarty PK, Fleites LA, Rayside PA, Hopkins DL, Gabriel DW. Three new Pierce’s disease pathogenicity effectors identified using Xylella fastidiosa biocontrol strain EB92-1. PLoS One. 2015;10:e0133796
Gouran H, Gillespie H, Nascimento R, Chakraborty S, Zaini PA, Jacobson A, et al. The secreted protease PrtA controls cell. Growth, Biofilm Form Pathog Xylella Fasti Sci Rep. 2016;6:31098
Carroll RK, Rivera FE, Cavaco CK, Johnson GM, Martin D, Shaw LN. The lone S41 family C-terminal processing protease in Staphylococcus aureus is localized to the cell wall and contributes to virulence. Microbiol-Sgm. 2014;160:1737–48.
Denance N, Legendre B, Briand M, Olivier V, de Boisseson C, Poliakoff F, et al. Several subspecies and sequence types are associated with the emergence of Xylella fastidiosa in natural settings in France. Plant Pathol. 2017;66:1054–64.
Fraser C, Hanage WP, Spratt BG. Recombination and the nature of bacterial speciation. Science. 2007;315:476–80.
Coletta HD, Francisco CS, Lopes JRS, Muller C, Almeida RPP. Homologous recombination and Xylella fastidiosa host-pathogen associations in South America. Phytopathology. 2017;107:305–12.
Nunney L, Ortiz B, Russell SA, Sanchez RR, Stouthamer R. The complex biogeography of the plant pathogen Xylella fastidiosa: genetic evidence of introductions and subspecific introgression in Central America. PLoS One. 2014;9:e112463.
Jacques MA, Denance N, Legendre B, Morel E, Briand M, Mississipi S, et al. New coffee plant-infecting Xylella fastidiosa variants derived via homologous recombination. Appl Environ Microbiol. 2016;82:1556–68.
Mostowy R, Croucher NJ, Hanage WP, Harris SR, Bentley S, Fraser C. Heterogeneity in the frequency and characteristics of homologous recombination in pneumococcal evolution. Plos Genetics. 2014;10:e1004300.
Giampetruzzi A, Loconsole G, Boscia D, Calzolari A, Chiumenti M, Martelli GP, et al. Draft genome sequence of CO33, a coffee-infecting isolate of Xylella fastidiosa. Genome Announcements. 2015;3:e01472-15.
Kloesges T, Popa O, Martin W, Dagan T. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol. 2011;28:1057–74.
Arnold BJ, Gutmann MU, Grad YH, Sheppard SK, Corander J, Lipsitch M, et al. Weak epistasis may drive adaptation in recombining bacteria. Genetics. 2018;208:1247–60.
Chatterjee S, Almeida RPP, Lindow S. Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu Rev Phytopathol. 2008;46:243–71.
De La Fuente L, Montanes E, Meng YZ, Li YX, Burr TJ, Hoch HC, et al. Assessing adhesion forces of type I and type IV pili of Xylella fastidiosa bacteria by use of a microfluidic flow chamber. Appl Environ Microbiol. 2007;73:2690–6.
Feil H, Feil WS, Lindow SE. Contribution of fimbrial and afimbrial adhesins of Xylella fastidiosa to attachment to surfaces and virulence to grape. Phytopathology. 2007;97:318–24.
Killiny N, Almeida RPP. Factors affecting the initial adhesion and retention of the plant pathogen Xylella fastidiosa in the foregut of an insect vector. Appl Environ Microbiol. 2014;80:420–6.
Meng YZ, Li YX, Galvani CD, Hao GX, Turner JN, Burr TJ, et al. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J Bacteriol. 2005;187:5560–7.
Roper MC, Greve LC, Warren JG, Labavitch JM, Kirkpatrick BC. Xylella fastidiosa requires polygalacturonase for colonization and pathogenicity in Vitis vinifera grapevines. Mol Plant-Microbe Interact. 2007;20:411–9.
Rapicavoli JN, Blanco-Ulate B, Muszyński A, Figueroa-Balderas R, Morales-Cruz A, Azadi P. et al. Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat Commun. 2018;9:390
Killiny N, Almeida RPP. Gene regulation mediates host specificity of a bacterial pathogen. Environ Microbiol Rep. 2011;3:791–7.
Pierce BK, Kirkpatrick BC. The PhoP/Q two-component regulatory system is essential for Xylella fastidiosa survival in Vitis vinifera grapevines. Physiol Mol Plant Pathol. 2015;89:55–61.
Rosnow JJ, Hwang S, Killinger BJ, Kim YM, Moore RJ, Lindemann SR, et al. A cobalamin activity-based probe enables microbial cell growth and finds new cobalamin-protein interactions across domains. Appl Environ Microbiol. 2018;84:e00955–18.
Cordonnier C, Le Bihan G, Emond-Rheault J-G, Garrivier A, Harel J, Jubelin G. Vitamin B12 uptake by the gut commensal bacteria Bacteroides thetaiotaomicron limits the production of shiga toxin by enterohemorrhagic Escherichia coli. Toxins. 2016;8:14
Lee K-M, Go J, Yoon MY, Park Y, Kim SC, Yong DE, et al. Vitamin B12-mediated restoration of defective anaerobic growth leads to reduced biofilm formation in Pseudomonas aeruginosa. Infect Immun. 2012;80:1639–49.
Bjork GR, Ericson JU, Gustafsson CED, Hagervall TG, Jonsson YH, Wikstrom PM. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–87.
Newman KL, Almeida RP, Purcell AH, Lindow SE. Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl Environ Microbiol. 2003;69:7319–27.
Matsumoto A, Young GM, Igo MM. Chromosome-based genetic complementation system for Xylella fastidiosa. Appl Environ Microbiol. 2009;75:1679–87.
Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–47.
Kavita K, de Mets F, Gottesman S. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr Opin Microbiol. 2018;42:53–61.
O’Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res. 2015;277:32–48.
De La Fuente L, Parker JK, van Santen E, Oliver JE, Brannen P, Granger S, et al. The bacterial pathogen Xylella fastidiosa affects the leaf ionome of plant hosts during infection. PLoS One. 2013;8:e62945
Rock CD. Trans-acting small interfering RNA4: key to nutraceutical synthesis in grape development?. Trends Plant Sci. 2013;18:601–10.
Antolin-Llovera M, Ried MK, Binder A, Parniske M. Receptor kinase signaling pathways in plant-microbe interactions. In: VanAlfen NK, Leach JE, Lindow S, eds. Annual review of phytopathology, Vol 50. Palo Alto: Annual Reviews; 2012. p. 451–73.
Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–36.
Hu Z, Dong HJ, Ma JC, Yu YH, Li KH, Guo QQ, et al. Novel Xanthomonas campestris long-chain-specific 3-oxoacyl-acyl carrier protein reductase involved in diffusible signal factor synthesis. Mbio. 2018;9:e00596–18.
Mao YH, Li F, Ma JC, Hu Z, Wang HH. Sinorhizobium meliloti functionally replaces 3-oxoacyl-acyl carrier protein reductase (FabG) by overexpressing NodG during fatty acid synthesis. Mol Plant-Microbe Interact. 2016;29:458–67.
Filip’echeva YA, Shelud’ko AV, Prilipov AG, Burygin GL, Telesheva EM, Yevstigneyeva SS, et al. Plasmid AZOBR_p1-borne fabG gene for putative 3-oxoacyl-acyl-carrier protein reductase is essential for proper assembly and work of the dual flagellar system in the alphaproteobacterium Azospirillum brasilense Sp245. Can J Microbiol. 2018;64:107–18.
Selkrig J, Belousoff MJ, Headey SJ, Heinz E, Shiota T, Shen HH, et al. Conserved features in TamA enable interaction with TamB to drive the activity of the translocation and assembly module. Sci Rep. 2015;5:12905.
Acknowledgements
This project was funded by Agriculture and Food Research Initiative competitive grant no. 2015-67014-23085 from the USDA National Institute of Food and Agriculture, and HATCH AAES (Alabama Agricultural Experiment Station) program provided to N.P and L.D, and partly supported by research grant OS 2015330 of the Ministry of Economic Affairs in the Netherlands. We thank the Alabama Supercomputing Authority for granting access to their high-performance computing platform.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Potnis, N., Kandel, P.P., Merfa, M.V. et al. Patterns of inter- and intrasubspecific homologous recombination inform eco-evolutionary dynamics of Xylella fastidiosa. ISME J 13, 2319–2333 (2019). https://doi.org/10.1038/s41396-019-0423-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-019-0423-y
This article is cited by
-
Botanical products for managing Philaenus spumarius, vector of Xylella fastidiosa subspecies pauca
Journal of Pest Science (2025)
-
Impacts of local population history and ecology on the evolution of a globally dispersed pathogen
BMC Genomics (2020)
-
Citrus Variegated Chlorosis: an Overview of 30 Years of Research and Disease Management
Tropical Plant Pathology (2020)