Abstract
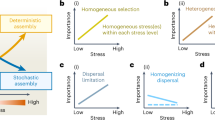
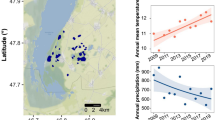
Similarities and differences of phenotypes within local co-occurring species hold the key to inferring the contribution of stochastic or deterministic processes in community assembly. Developing both phylogenetic-based and trait-based quantitative methods to unravel these processes is a major aim in community ecology. We developed a trait-based approach that: (i) assesses if a community trait clustering pattern is related to increasing environmental constraints along a gradient; and (ii) determines quantitative thresholds for an environmental variable along a gradient to interpret changes in prevailing community assembly drivers. We used a regional set of natural shallow saline ponds covering a wide salinity gradient (0.1–40% w/v). We identify a consistent discrete salinity threshold (ca. 5%) for microbial community assembly drivers. Above 5% salinity a strong environmental filtering prevailed as an assembly force, whereas a combination of biotic and abiotic factors dominated at lower salinities. This method provides a conceptual approach to identify consistent environmental thresholds in community assembly and enables quantitative predictions for the ecological impact of environmental changes.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
van der Plas F, Janzen T, Ordóñez A, Fokema W, Reinders J, Etienne RS, et al. A new modeling approach estimates the relative importance of different community assembly processes. Ecology. 2015;96:1502–15.
Vellend M. Conceptual synthesis in community ecology. Q Rev Biol. 2010;85:183–206.
Vellend M. The theory of ecological communities (MPB-57). Princeton University Press; 2016.
Gravel D, Canham CD, Beaudet M, Messier C. Reconciling niche and neutrality: the continuum hypothesis. Ecol Lett. 2006;9:399–409.
MacArthur RH. Species packing and what interspecific competition minimizes. Proc Natl Acad Sci USA. 1969;64:1369–71.
Hubbell SP. The unified theory of biodiversity and biogeography. Princeton: Princeton University Press; 2001.
Tilman D. Resource competition and community structure. Princeton, NJ: Princeton University Press; 1982.
Law R, Morton RD. Permanence and the assembly of ecological communities. Ecology. 1996;77:762–75.
Law R, Weatherby AJ, Warren PH. On the invasibility of persistent protist communities. Oikos. 2000;88:319–26.
Mayfield MM, Levine JM. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol Lett. 2010;13:1085–93.
Chesson PL. Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst. 2000;31:343–66.
Liebhold AM, Chase J. Metacommunity ecology. Princeton: Princeton University Press; 2018.
Bertness M, Callaway RM. Positive interactions in communities. Trends Ecol Evol. 1994;9:191–3.
Maestre FT, Callaway RM, Valladares F, Lortie CJ. Refining the stress-gradient hypothesis for competition and facilitation in plant communities. J Ecol. 2009;97:199–205.
Bastolla U, Fortuna MA, Pascual-García A, Ferrera A, Luque B, Bascompte J. The architecture of 290 mutualistic networks minimizes competition and increases biodiversity. Nature. 2009;458:1018–20.
Helmus MR, Savage K, Diebel MW, Maxted JT, Ives AR. Separating the determinants of phylogenetic community structure. Ecol Lett. 2007;10:917–25.
Kraft NJB, Adler PB, Godoy O, James EC, Fuller S, Levine JM. Community assembly, coexistence and the environmental filtering metaphor. Funct Ecol. 2014;29:592–9.
Peres-Neto PR, Dray S, Braak CJ. Linking trait variation to the environment: critical issues with community-weighted mean correlation resolved by the fourth-corner approach. Ecography. 2017;40:806–16. arXiv:arXiv:0902.0132v1
Wisz MS, Pottier J, Kissling WD, Pellissier L, Lenoir J, Damgaard CF, et al. The role of biotic interactions in shaping distributions and realised assemblages of species: implications for species distribution modelling. Biol Rev. 2013;88:15–30.
Townsend Peterson A, Soberón J, Pearson RG, Anderson RP, Martínez-Meyer E, Nakamura M, et al. Ecological niches and geographic distributions. Oxford: Princeton University Press; 2011.
Barberán A, Casamayor EO, Fierer N. The microbial contribution to macroecology. Front Microbiol. 2014;5:203.
Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci USA. 2015;112:E1326–E1332.
Goberna M, Navarro-Cano JA, Valiente-Banuet A, García C, Verdú M. Abiotic stress tolerance and competition-related traits underlie phylogenetic clustering in soil bacterial communities. Ecol Lett. 2014;17:1191–201.
Barberán A, Ramirez KS, Leff JW, Bradford MA, Wall DH, Fierer N. Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol Lett. 2014;17:794–802.
Ter Braak CJ, Cormont A, Dray S. In the fourth-corner problem reports. Ecology. 2012;93:1525–6.
Edwards KF, Litchman E, Klausmeier CA. Functional traits explain phytoplankton responses to environ mental gradients across lakes of the United States. Ecology. 2013;94:1626–35.
Etienne RS, Alonso D. A dispersal-limited sampling theory for species and alleles. Ecol Lett. 2005;8:1147–56. arXiv.
Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu Rev Ecol Systemat. 2002;33:475–505.
Gotelli NJ, Graves GR. Null models in ecology. Washington: Smithsonian Institution Press; 1996.
Casamayor EO, Triadó-Margarit X, Castañeda C. Microbial biodiversity in saline shallow lakes of the Monegros Desert, Spain. FEMS Microbiol Ecol. 2013;85:503–18.
Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8.
Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:581–91.
Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Pillay M, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42:D560–7.
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnol. 2013;31:814–21.
Nemergut DR, Knelman JE, Ferrenberg S, Bilinski T, Melbourne B, Jiang L, et al. Decreases in average bacterial community rRNA operon copy number during succession. ISME J. 2015;10:1147–56.
Zeileis A, Kleiber C, Krämer W, Hornik K. Testing and dating of structural changes in practice. Comput Stat Data Anal. 2003;44:109–23.
McGill BJ, Enquist BJ, Weiher E, Westoby M. Rebuilding community ecology from functional traits. Trends Ecol Evol. 2006;21:178–85.
Adler PB, Fajardo A, Kleinhesselink AR, Kraft NJB. Trait-based tests of coexistence mechanisms. Ecol Lett. 2013;16:1294–306.
Thakur MP, Wright AJ. Environmental filtering, niche construction, and trait variability: the missing discussion. Trends Ecol Evol. 2017;32:884–6.
Barberán A, Fernández-Guerra A, Bohannan BJM, Casamayor EO. Exploration of community traits as ecological markers in microbial metagenomes. Mol Ecol. 2012;21:1909–17.
Krause S, Le Roux X, Niklaus PA, Van Bodegom PM, Lennon JT, Bertilsson S, et al. Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front Microbiol. 2014;5:251.
Ortiz-Álvarez R, Fierer N, de los Ríos A, Casamayor EO, Barberán A. Consistent changes in the taxonomic structure and functional attributes of bacterial communities during primary succession. ISME J. 2018;12:1658–67.
Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci. 2011;108:14288–93.
Grossmann L, Beisser D, Bock C, Chatzinotas A, Jensen M, Preisfeld A, et al. Trade-off between taxon diversity and functional diversity in European lake ecosystems. Mol Ecol. 2016;25:5876–88.
Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O’Connor MI, et al. Function and functional redundancy in microbial systems. Nat Ecol Evol. 2018;2:936–43.
von Heijne G. Signal sequences: the limits of variation. J Mol Biol. 1985;184:99–105.
Wesley RR, Brüser T, Kissinger JC, Pohlschröder M. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol. 2002;45:943–50.
van der Ploeg R, Mäder U, Homuth G, Schaffer M, Denham EL, Monteferrante CG, et al. Environmental salinity determines the specificity and need for tat-dependent secretion of the YwbN protein in Bacillus subtilis. PLoS One. 2011;6:1–10.
van der Ploeg R, Monteferrante CG, Piersma S, Barnett JP, Kouwen TRHM, Robinson C, et al. High-salinity growth conditions promote tat-independent secretion of tat substrates in Bacillus subtilis. Appl Environ Microbiol. 2012;78:7733–44.
Paul S, Bag SK, Das S, Harvill ET, Dutta C. Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 2008;9:R70.
Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–33.
Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–40.
Auguet J-C, Barberán A, Casamayor EO. Global ecological patterns in uncultured Archaea. ISME J. 2010;4:182–90.
Barberán A, Casamayor EO. Global phylogenetic community structure and beta-diversity patterns in surface bacterioplankton metacommunities. Aquat Microb Ecol. 2010;59:1–10.
Gasol JM, Casamayor EO, Joint I, Garde K, Gustavson K, Benlloch S, et al. Control of heterotrophic prokaryotic abundance and growth rate in hypersaline planktonic environments. Aquat Microb Ecol. 2004;34:193–206.
Casamayor EO, Triadó-Margarit X. Microbial diversity and novelty along salinity gradients. New York: Springer; 2013. p. 1–8.
Oren A. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev. 1999;63:334–48.
Bunin G. Ecological communities with Lotka–Volterra dynamics. Phys Rev E. 2017;95:1–8.
Acknowledgements
This work was funded by the Spanish "Ministerio de Economía y Competitividad" under the projects BRIDGES (CGL2015-69043-P, to EOC and DA), and the Ramón y Cajal Fellowship program (DA). Both VJO and MM-S have been supported by Ph.D. contracts funded by the Spanish "Ministerio de Economía y Competitividad" under the projects BRIDGES and SITES (CGL2015-69043-P, CGL2012-39964 to EOC and DA). We thank Joan Cáliz and Gerard Funosas for help in bioinformatic analyses and fruitful discussions and the facilities and warm and creative atmosphere provided by the "white room" in the Computational Biology Lab (CBL) of the Center for Advanced Studies of Blanes.
Author information
Authors and Affiliations
Contributions
EOC, JAC, and DA conceived the study. EOC and XT-M carried out the sampling and field work. JAC, VJO and DA designed the null models. RO-A, MM-S, and XT-M conducted bioinformatic analyses. MM-S, VJO, JAC, and XT-M contributed with scripts and analyses. JAC and XT-M led the analyses of the data. XT-M, JAC, and MM-S assembled and organized the supplementary materials. XT-M, JAC, and DA led the writing of the manuscript. All authors contributed to developing the concepts along several “Bridges” workshops, discussed and contributed critically to the drafts and gave final approval for publication.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Triadó-Margarit, X., Capitán, J.A., Menéndez-Serra, M. et al. A Randomized Trait Community Clustering approach to unveil consistent environmental thresholds in community assembly. ISME J 13, 2681–2689 (2019). https://doi.org/10.1038/s41396-019-0454-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-019-0454-4
This article is cited by
-
Selection dictates the distance pattern of similarity in trees and soil fungi across forest ecosystems
Fungal Diversity (2024)
-
Ecological and Metabolic Thresholds in the Bacterial, Protist, and Fungal Microbiome of Ephemeral Saline Lakes (Monegros Desert, Spain)
Microbial Ecology (2021)
-
Towards sustainable agriculture: rhizosphere microbiome engineering
Applied Microbiology and Biotechnology (2021)