Abstract
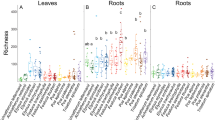
Specialized associations between interacting species fundamentally determine the diversity and distribution of both partners. How the specialization of guilds of organisms varies along environmental gradients underpins popular theories of biogeography and macroecology, whereas the degree of specialization of a species is typically considered fixed. However, the extent to which environmental context impacts specialization dynamics is seldom examined empirically. In this study, we examine how specialization within a bipartite network consisting of three co-occurring plant species and their foliar fungal endophyte symbionts changes along a 1000-meter elevation gradient where host species were held constant. The gradient, along the slope of Mauna Loa shield volcano, represents almost the entire elevational range of two of the three plants. Network and plant specialization values displayed a parabolic relationship with elevation, and were highest at middle elevations, whereas bipartite associations were least specific at low and high elevations. Shannon’s diversity of fungal endophytes correlated negatively with specificity, and was highest at the ends of the transects. Although plant host was a strong determinant of fungal community composition within sites, fungal species turnover was high among sites. There was no evidence of spatial or elevational patterning in fungal community compositon. Our work demonstrates that specificity can be a plastic trait, which is influenced by the environment and centrality of the host within its natural range.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Hynson NA, Bruns TD. Fungal hosts for mycoheterotrophic plants: a nonexclusive, but highly selective club. New Phytol. 2010;185:598–601.
Bohlool BB, Schmidt EL. Lectins: a possible basis for specificity in the Rhizobium—legume root nodule symbiosis. Science. 1974;185:269–71.
Grubisha LC, Trappe JM, Molina R, Spatafora JW. Biology of the ectomycorrhizal genus Rhizopogon. VI. Re-examination of infrageneric relationships inferred from phylogenetic analyses of ITS sequences. Mycologia. 2002;94:607–19.
Bruns TD, Bidartondo MI, Taylor DL. Host specificity in ectomycorrhizal communities: what do the exceptions tell us? Integr Comp Biol. 2002;42:352–9.
Klironomos JN. Host-specificity and functional diversity among arbuscular mycorrhizal fungi. Micro Biosyst New Front. 2000;1:845–51.
Smith SE, Read DJ. Mycorrhizal symbiosis. 3rd ed. London, UK: Academic Press; 2008.
Gilbert GS, Webb CO. Phylogenetic signal in plant pathogen-host range. Proc Natl Acad Sci USA. 2007;104:4979–83.
Blüthgen N, Menzel F, Blüthgen N. Measuring specialization in species interaction networks. BMC Ecol. 2006;6:9.
Mariadassou M, Pichon S, Ebert D. Microbial ecosystems are dominated by specialist taxa. Ecol Lett. 2015;18:974–82.
Waring B, Hawkes CV. Ecological mechanisms underlying soil bacterial responses to rainfall along a steep natural precipitation gradient. FEMS Microbiol Ecol. 2018;94:1–10.
Connell J. On the role of natural enemies in preventing competitive exclusion in some marine animals and in rain forest trees. Dyn Numbers Popul Proc Adv Study Inst Dyn Numbers Popul Oosterbeck. 1971;298–312.
Janzen DH. Herbivores and the number of tree species in tropical forests. Am Nat. 1970;104:501–28.
Forister ML, Novotny V, Panorska AK, Baje L, Basset Y, Butterill PT, et al. The global distribution of diet breadth in insect herbivores. Proc Natl Acad Sci USA. 2015;112:442–7.
Schemske DW, Mittelbach GG, Cornell HV, Sobel JM, Roy K. Is there a latitudinal gradient in the importance of biotic interactions? Annu Rev Ecol Evol Syst. 2009;40:245–69.
Poisot T, Stouffer DB, Gravel D. Beyond species: why ecological interaction networks vary through space and time. Oikos. 2015;124:243–51.
Körner C. The use of ‘altitude’ in ecological research. Trends Ecol Evol. 2007;22:569–74.
Wagner WL. Manual of the flowering plants of Hawaiʻi, Rev. ed. Honolulu, HI: University of Hawaiʻi Press: Bishop Museum Press; 1999.
Vitousek PM, Matson PA, Turner DR. Elevational and age gradients in Hawaiian montane rainforest: foliar and soil nutrients. Oecologia. 1988;77:565–70.
Stone JK, Bacon CW, White Jr JF. Endophytism defined. In: Bacon CW, White Jr JF, editors. An overview of endophytic microbes. New York, NY: Marcel Dekker, Inc.; 2000. p. 3–29.
Arnold AE. Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev. 2007;21:51–66.
Arnold AE, Engelbrecht BMJ. Fungal endophytes nearly double minimum leaf conductance in seedlings of a neotropical tree species. J Trop Ecol. 2007;23:369–72.
Kannadan S, Rudgers JA. Endophyte symbiosis benefits a rare grass under low water availability. Funct Ecol. 2008;22:706–13.
Zahn G, Amend AS. Foliar microbiome transplants confer disease resistance in a critically-endangered plant. PeerJ. 2017;5:e4020.
Arnold AE, Herre EA. Canopy cover and leaf age affect colonization by tropical fungal endophytes: ecological pattern and process in Theobroma cacao (Malvaceae). Mycologia. 2003;95:388–98.
U’Ren JM, Lutzoni F, Miadlikowska J, Laetsch AD, Arnold AE. Host and geographic structure of endophytic and endolichenic fungi at a continental scale. Am J Bot. 2012;99:898–914.
Vincent JB, Weiblen GD, May G. Host associations and beta diversity of fungal endophyte communities in New Guinea rainforest trees. Mol Ecol. 2016;25:825–41.
Bálint M, Tiffin P, Hallström B, O’Hara RB, Olson MS, Fankhauser JD, et al. Host genotype shapes the foliar fungal microbiome of balsam poplar (Populus balsamifera). PLoS ONE. 2013;8:e53987.
Saunders M, Kohn LM. Evidence for alteration of fungal endophyte community assembly by host defense compounds. New Phytol. 2009;182:229–38.
Schleuning M, Fründ J, Klein A-M, Abrahamczyk S, Alarcón R, Albrecht M, et al. Specialization of mutualistic interaction networks decreases toward tropical latitudes. Curr Biol. 2012;22:1925–31.
Smith DP, Peay KG. Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS ONE. 2014;9:e90234.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41.
Nguyen NH, Smith D, Peay K, Kennedy P. Parsing ecological signal from noise in next generation amplicon sequencing. New Phytol. 2015;205:1389–93.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1.
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–7.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Team RC. R language definition. Vienna Austria R Found Stat Comput 2000.
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: community ecology package; 2017.
Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27:325–49.
Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209.
Baselga A. Partitioning abundance-based multiple-site dissimilarity into components: balanced variation in abundance and abundance gradients. Methods Ecol Evol. 2017;8:799–808.
Baselga A, Orme CDL. betapart: an R package for the study of beta diversity: Betapart package. Methods Ecol Evol. 2012;3:808–12.
Anderson MJ. A new method for non-parametric multivariate analysis of variance: non-parametric manova for ecology. Austral Ecol. 2001;26:32–46.
Dormann CF, Gruber B, Fründ J. Introducing the bipartite package: analysing ecological networks. Interaction. 2008;1:0–2413793.
Peña EA, Slate EH. Global validation of linear model assumptions. J Am Stat Assoc. 2006;101:341–54.
Cobian G, Amend A, Egan C. Plant–microbe specificity varies as a function of elevation. Figshare 2019.
Parker IM, Saunders M, Bontrager M, Weitz AP, Hendricks R, Magarey R, et al. Phylogenetic structure and host abundance drive disease pressure in communities. Nature. 2015;520:542–4.
Schirrmann MK, Zoller S, Croll D, Stukenbrock EH, Leuchtmann A, Fior S. Genomewide signatures of selection in Epichloë reveal candidate genes for host specialization. Mol Ecol. 2018;27:3070–86.
Smith GR, Steidinger BS, Bruns TD, Peay KG. Competition–colonization tradeoffs structure fungal diversity. ISME J. 2018;12:1758–67.
Brown JH. On the relationship between abundance and distribution of species. Am Nat. 1984;124:255–79.
Caughley G, Grice D, Barker R, Brown B. The edge of the range. J Anim Ecol. 1988;57:771.
Jump AS, Woodward FI, Burke T. Cirsium species show disparity in patterns of genetic variation at their range-edge, despite similar patterns of reproduction and isolation. New Phytol. 2003;160:359–70.
Sorte C, Hofmann G. Changes in latitudes, changes in aptitudes: Nucella canaliculata (Mollusca: Gastropoda) is more stressed at its range edge. Mar Ecol Prog Ser. 2004;274:263–8.
Lipp CC, Goldstein G, Meinzer FC, Niemczura W. Freezing tolerance and avoidance in high-elevation Hawaiian plants. Plant Cell Environ. 1994;17:1035–44.
Vander Kloet SP, Avery TS. Vaccinium on the edge. Edinb J Bot. 2010;67:7–24.
Rauw WM. Immune response from a resource allocation perspective. Front Genet. 2012;3:1–14.
Lau MK, Arnold AE, Johnson NC. Factors influencing communities of foliar fungal endophytes in riparian woody plants. Fungal Ecol. 2013;6:365–78.
David AS, Seabloom EW, May G. Plant host species and geographic distance affect the structure of aboveground fungal symbiont communities, and environmental filtering affects belowground communities in a coastal dune ecosystem. Microb Ecol. 2016;71:912–26.
Lamit LJ, Lau MK, Sthultz CM, Wooley SC, Whitham TG, Gehring CA. Tree genotype and genetically based growth traits structure twig endophyte communities. Am J Bot. 2014;101:467–78.
Giauque H, Hawkes CV. Climate affects symbiotic fungal endophyte diversity and performance. Am J Bot. 2013;100:1435–44.
Zimmerman NB, Vitousek PM. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc Natl Acad Sci USA. 2012;109:13022–7.
Darcy JL, Cobian GM, Swift SOI, Zahn GL, Perry BA, Amend AS. Fungal communities living within leaves of native Hawaiian dicots are structured by landscape-scale variables as well as by host plants. bioRxiv 2019;640029.
Miyamoto Y, Nakano T, Hattori M, Nara K. The mid-domain effect in ectomycorrhizal fungi: range overlap along an elevation gradient on Mount Fuji, Japan. ISME J. 2014;8:1739–46.
Singh D, Takahashi K, Kim M, Chun J, Adams JM. A hump-backed trend in bacterial diversity with elevation on Mount Fuji, Japan. Microb Ecol. 2012;63:429–37.
Fierer N, McCain CM, Meir P, Zimmermann M, Rapp JM, Silman MR, et al. Microbes do not follow the elevational diversity patterns of plants and animals. Ecology. 2011;92:797–804.
Bryant JA, Lamanna C, Morlon H, Kerkhoff AJ, Enquist BJ, Green JL. Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc Natl Acad Sci USA. 2008;105:11505–11.
Egan CP, Callaway RM, Hart MM, Pither J, Klironomos J. Phylogenetic structure of arbuscular mycorrhizal fungal communities along an elevation gradient. Mycorrhiza. 2017;27:273–82.
Acknowledgements
The authors wish to thank Erin Datlof at UH Hilo and Tomoko Sakishima at UNLV for field assistance, Nhu Nguyen for bioinformatics assistance, and members of the Amend and Hynson labs for critical review of earlier versions of the manuscript. We appreciate Dr. Heather Sahli for contributing data on plant cover. The authors also wish to thank the National Science Foundation for their generous support as NSF grant #1255972 to ASA and NSF grant #1329626 to GMC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cobian, G.M., Egan, C.P. & Amend, A.S. Plant–microbe specificity varies as a function of elevation. ISME J 13, 2778–2788 (2019). https://doi.org/10.1038/s41396-019-0470-4
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41396-019-0470-4
This article is cited by
-
Interplay of biotic and abiotic factors shapes tree seedling growth and root-associated microbial communities
Communications Biology (2024)
-
Microbiome specificity and fluxes between two distant plant taxa in Iberian forests
Environmental Microbiome (2023)
-
Plant and fungal species interactions differ between aboveground and belowground habitats in mountain forests of eastern China
Science China Life Sciences (2023)
-
Plant-microbe interactions in the phyllosphere: facing challenges of the anthropocene
The ISME Journal (2022)
-
Grass species identity shapes communities of root and leaf fungi more than elevation
ISME Communications (2022)