Abstract
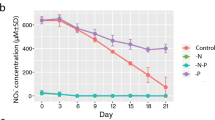
Phytoplankton face environmental nutrient variations that occur in the dynamic upper layers of the ocean. Phytoplankton cells are able to rapidly acclimate to nutrient fluctuations by adjusting their nutrient-uptake system and metabolism. Disentangling these acclimation responses is a critical step in bridging the gap between phytoplankton cellular physiology and community ecology. Here, we analyzed the dynamics of phosphate (P) uptake acclimation responses along different P temporal gradients by using batch cultures of the diatom Phaeodactylum tricornutum. We employed a multidisciplinary approach that combined nutrient-uptake bioassays, transcriptomic analysis, and mathematical models. Our results indicated that cells increase their maximum nutrient-uptake rate (Vmax) both in response to P pulses and strong phosphorus limitation. The upregulation of three genes coding for different P transporters in cells experiencing low intracellular phosphorus levels supported some of the observed Vmax variations. In addition, our mathematical model reproduced the empirical Vmax patterns by including two types of P transporters upregulated at medium-high environmental and low intracellular phosphorus levels, respectively. Our results highlight the existence of a sequence of acclimation stages along the phosphate continuum that can be understood as a succession of acclimation responses. We provide a novel conceptual framework that can contribute to integrating and understanding the dynamics and wide diversity of acclimation responses developed by phytoplankton.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Falkowski PG, Barber RT, Smetacek V. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281:200–6.
Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–40.
Reynolds CS. The ecology of phytoplankton. Cambridge University Press; Cambridge, UK: 2006.
Reynolds C, Dokulil M, Padisák J. Understanding the assembly of phytoplankton in relation to the trophic spectrum: where are we now? Hydrobiologia. 2000;424:147–52.
Caperon J, Meyer J. Nitrogen-limited growth of marine phytoplankton—II. Uptake kinetics and their role in nutrient limited growth of phytoplankton. Deep-Sea Res. 1972;19:619–32.
Gotham IJ, Rhee G-Y. Comparative kinetic studies of phosphate‐limited growth and phosphate uptake in phytoplankton in continuous culture. J Phycol. 1981;17:257–65.
Morel FMM. Kinetics of nutrient uptake and growth in phytoplankton. J Phycol. 1987;23:137–50.
Zhang S-F, Yuan C-J, Chen Y, Chen X-H, Li D-X, Liu J-L, et al. Comparative transcriptomic analysis reveals novel insights into the adaptive response of Skeletonema costatum to changing ambient phosphorus. Front Microbiol. 2016;7:1476.
Krumhardt KM, Callnan K, Roache-Johnson K, Swett T, Robinson D, Reistetter EN, et al. Effects of phosphorus starvation versus limitation on the marine cyanobacterium Prochlorococcus MED4 I: uptake physiology. Environ Microbiol. 2013;15:2114–28.
Lomas MW, Bonachela JA, Levin SA, Martiny AC. Impact of ocean phytoplankton diversity on phosphate uptake. Proc Natl Acad Sci USA. 2014;111:17540–5.
Sunda WG, Huntsman SA. Regulation of cellular manganese and manganese transport rates in the unicellular alga Chlamydomonas. Limnol Oceano. 1985;30:71–80.
Riegman R, Stolte W, Noordeloos AAM, Slezak D. Nutrient uptake and alkaline phosphatase (EC 3: 1: 3: 1) activity of Emiliania huxleyi (Prymnesiophyceae) during growth under N and P limitation in continuous cultures. J Phycol. 2000;36:87–96.
Michaelis L, Menten ML. Die kinetik der invertinwirkung. Biochem Z. 1913;49:333–69.
Droop MR. Vitamin B12 and marine ecology. IV. The kinetics of uptake, growth and inhibition in Monochrysis lutheri. J Mar Biol Assoc U Kingd. 1968;48:689–733.
Aksnes DL, Egge JK. A theoretical model for nutrient uptake in phytoplankton. Mar Ecol Prog Ser. 1991;70:65–72.
Litchman E, Klausmeier CA. Trait-based community ecology of phytoplankton. Annu Rev Ecol Evol Syst. 2008;39:615–39.
Reistetter EN, Krumhardt K, Callnan K, Roache-Johnson K, Saunders JK, Moore LR, et al. Effects of phosphorus starvation versus limitation on the marine cyanobacterium Prochlorococcus MED4 II: gene expression. Environ Microbiol. 2013;15:2129–43.
Healey FP. Slope of the Monod equation as an indicator of advantage in nutrient competition. Micro Ecol. 1980;5:281–6.
Fu F, Zhang Y, Bell PRF, Hutchins DA. Phosphate uptake and growth kinetics of Trichodesmium (Cyanobacteria) isolates from the North Atlantic Ocean and the Great Barrier Reef, Australia. J Phycol. 2005;41:62–73.
Collos Y, Vaquer A, Souchu P. Acclimation of nitrate uptake by phytoplankton to high substrate levels. J Phycol 2005;41:466–78.
Rivkin RB, Swift E. Phosphate uptake by the oceanic dinoflagellate Pyrocystis noctiluca. J Phycol. 1982;18:113–20.
Spijkerman E, Coesel PFM. Phosphorus uptake and growth kinetics of two planktonic desmid species. Eur J Phycol. 1996;31:53–60.
Lin S, Litaker RW, Sunda WG. Phosphorus physiological ecology and molecular mechanisms in marine phytoplankton. J Phycol. 2016;52:10–36.
Martiny AC, Coleman ML, Chisholm SW. Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc Natl Acad Sci USA. 2006;103:12552–7.
Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–99.
Dyhrman ST, Jenkins BD, Rynearson TA, Saito MA, Mercier ML, Alexander H, et al. The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS ONE. 2012;7:e33768.
Liu Z, Koid AE, Terrado R, Campbell V, Caron DA, Heidelberg KB. Changes in gene expression of Prymnesium parvum induced by nitrogen and phosphorus limitation. Front Microbiol. 2015;6:631.
Cruz de Carvalho MH, Sun H, Bowler C, Chua N. Noncoding and coding transcriptome responses of a marine diatom to phosphate fluctuations. New Phytol. 2016;210:497–510.
Smith SL, Yamanaka Y, Pahlow M, Oschlies A. Optimal uptake kinetics: physiological acclimation explains the pattern of nitrate uptake by phytoplankton in the ocean. Mar Ecol Prog Ser. 2009;384:1–12.
Dick GJ. Embracing the mantra of modellers and synthesizing omics, experiments and models. Environ Microbiol Rep. 2017;9:18–20.
Bonachela JA, Raghib M, Levin SA. Dynamic model of flexible phytoplankton nutrient uptake. Proc Natl Acad Sci USA. 2011;108:20633–8.
Button DK. Nutrient uptake by microorganisms according to kinetic parameters from theory as related to cytoarchitecture. Microbiol Mol Biol Rev. 1998;62:636–45.
Mann KH, Lazier JRN. Dynamics of marine ecosystems: biological-physical interactions in the oceans. John Wiley & Sons; Oxford, UK; 2013.
Visser M, Batten S, Becker G, Bot P, Colijn F, Damm P, et al. Time series analysis of monthly mean data of temperature, salinity, nutrients, suspended matter, phyto-and zooplankton at eight locations on the Northwest European shelf. Dtsch Hydrogr Z. 1996;48:299–323.
Blauw AN, Beninca E, Laane RWPM, Greenwood N, Huisman J. Dancing with the tides: fluctuations of coastal phytoplankton orchestrated by different oscillatory modes of the tidal cycle. PLoS One. 2012;7:e49319.
Tovar-Sanchez A, Sañudo-Wilhelmy SA, Garcia-Vargas M, Weaver RS, Popels LC, Hutchins DA. A trace metal clean reagent to remove surface-bound iron from marine phytoplankton. Mar Chem. 2003;82:91–9.
Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, Allen A, et al. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene. 2007;406:23–35.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:0034.1–0034.12.
Kaiserli E, Paldi K, O’Donnell L, Batalov O, Pedmale UV, Nusinow DA, et al. Integration of light and photoperiodic signaling in transcriptional nuclear foci. Dev Cell. 2015;35:311–21.
Quinn GP, Keough MJ. Experimental design and data analysis for biologists. Cambridge University Press; Cambridge, UK; 2002.
R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2017. http://www.R-project.org/
Pinheiro J, Bates D, DebRoy S, Sarkar D, Heisterkamp S, Van Willigen B, et al. Package “nlme.” Linear nonlinear Mix Eff Model. 2017; R package version 3.1-131. https://CRAN.R-project.org/package=nlme
Lenth R. Emmeans: estimated marginal means, aka least-squares means. 2018; R Package Version 1.3.0. https://CRAN.R-project.org/package=emmeans
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300.
Molina Grima E, García Camacho F, Sanchéz Pérez JA, Fernández Sevilla JM, Acién Fernández FG, Contreras Gómez A. A mathematical model of microalgal growth in light-limited chemostat culture. J Chem Technol Biotechnol Int Res Process Environ Clean Technol. 1994;61:167–73.
Olli K, Trunov K. Self-toxicity of Prymnesium parvum (Prymnesiophyceae). Phycologia. 2007;46:109–12.
Secco D, Wang C, Shou H, Whelan J. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 2012;586:289–95.
Dyhrman ST. Nutrients and their acquisition: phosphorus physiology in microalgae. In: The physiology of microalgae. MA Borowitzka, J Beardall, JA Raven, editors. Dordrecht, The Netherlands. Springer; 2016. p. 155–83.
Feng T-Y, Yang Z-K, Zheng J-W, Xie Y, Li D-W, Murugan SB, et al. Examination of metabolic responses to phosphorus limitation via proteomic analyses in the marine diatom Phaeodactylum tricornutum. Sci Rep. 2015;5:10373.
Hürlimann HC, Pinson B, Stadler‐Waibel M, Zeeman SC, Freimoser FM. The SPX domain of the yeast low‐affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep. 2009;10:1003–8.
Ghillebert R, Swinnen E, De Snijder P, Smets B, Winderickx J. Differential roles for the low-affinity phosphate transporters Pho87 and Pho90 in Saccharomyces cerevisiae. Biochem J. 2011;434:243–51.
Song B, Ward BB. Molecular cloning and characterization of high-affinity nitrate transporter in marine phytoplankton. J Phycol. 2007;43:542–52.
Menge DNL, Ballantyne F, Weitz JS. Dynamics of nutrient uptake strategies: lessons from the tortoise and the hare. Theor Ecol. 2011;4:163–77.
Perry MJ. Phosphate utilization by an oceanic diatom in phosphorus‐limited chemostat culture and in the oligotrophic waters of the central North Pacific. Limnol Oceano. 1976;21:88–107.
Klausmeier CA, Litchman E, Levin SA. A model of flexible uptake of two essential resources. J Theor Biol. 2007;246:278–89.
Zielinski BL, Allen AE, Carpenter EJ, Coles VJ, Crump BC, Doherty M, et al. Patterns of transcript abundance of eukaryotic biogeochemically-relevant genes in the Amazon River Plume. PLoS ONE. 2016;7:e0160929.
Toseland A, Daines SJ, Clark JR, Kirkham A, Strauss J, Uhlig C, et al. The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat Clim Chang. 2013;3:979–84.
Margalef R. Life-forms of phytoplankton as survival alternatives in an unstable environment. Oceano acta. 1978;1:493–509.
Acknowledgements
We thank the members of the Marine Resource Modeling group at Strathclyde University, specially M.R. Heath and M. Choua, for their comments on the model and our preliminary results. We would also like to acknowledge K. Griffiths for her help with 33P handling. M. W. Lomas kindly advised on the methodology for measuring P uptake. Suggestions and comments provided by James Grover and discussions with R. Anadón, F.G. Taboada, S. Romero-Romero and R. González-Gil are greatly appreciated. This research has been supported by the Marine Alliance for Science and Technology for Scotland (MASTS). C.C., S.S., and J.A.B. were supported by the Marine Alliance for Science and Technology for Scotland (MASTS) pooling initiative, funded by the Scottish Funding Council (HR09011) and contributing institutions. E.K. is grateful to the John Grieve Bequest and the Biotechnology and Biological Sciences Research Council (BBSRC) for the New Investigator Grant Award BB/M023079/1.
Funding
This research has been supported by the Marine Alliance for Science and Technology for Scotland (MASTS). C.C., S.S., and J.A.B. were supported by the Marine Alliance for Science and Technology for Scotland (MASTS) pooling initiative, funded by the Scottish Funding Council (HR09011) and contributing institutions. Gene expression experiments and analysis performed by E.K. were funded by the Biotechnology and Biological Sciences Research Council (BBSRC) (Grant Award BB/M023079/1).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cáceres, C., Spatharis, S., Kaiserli, E. et al. Temporal phosphate gradients reveal diverse acclimation responses in phytoplankton phosphate uptake. ISME J 13, 2834–2845 (2019). https://doi.org/10.1038/s41396-019-0473-1
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41396-019-0473-1
This article is cited by
-
Evaluating Potential Changes from Nitrogen to Phosphorus Nutrient Limitation of Phytoplankton Growth in North Inlet Estuary, SC
Estuaries and Coasts (2025)
-
Herbicide leakage into seawater impacts primary productivity and zooplankton globally
Nature Communications (2024)
-
Trypsin is a coordinate regulator of N and P nutrients in marine phytoplankton
Nature Communications (2022)
-
Phytoplankton and anthropogenic changes in pelagic environments
Hydrobiologia (2021)
-
Benthic and planktonic inorganic nutrient processing rates at the interface between a river and lake
Biogeochemistry (2021)