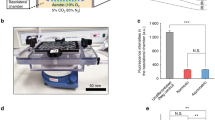
Abstract
Recent studies describe in detail the shifts in composition of human-associated polymicrobial communities from health to disease. However, the specific processes that drive the colonization and overgrowth of pathogens within these communities remain incompletely understood. We used in vitro culture systems and a disease-relevant mouse model to show that population size, which determines the availability of an endogenous diffusible small molecule, limits the growth, colonization, and in vivo virulence of the human oral pathogen Porphyromonas gingivalis. This bacterial pathogen overcomes the requirement for an endogenous cue by utilizing a cell-density dependent, growth-promoting, soluble molecule provided by the symbiotic early colonizer Veillonella parvula, but not produced by other commensals tested. Our work shows that exchange of cell-density-dependent diffusible cues between specific early and late colonizing species in a polymicrobial community drives microbial successions, pathogen colonization and disease development, representing a target process for manipulation of the microbiome towards the healthy state.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Miller MB, Bassler BL. Quorum sensing in bacteria. Ann Rev Microbiol. 2001;55:165–99.
Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nat. 2017;551:313–20.
Grossman AD. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Ann Rev Genet. 1995;29:477–508.
Kaprelyants AS, Kell DB. Do bacteria need to communicate with each other for growth? Trends Microbiol. 1996;4:237–42.
Mukamolova GV, Kaprelyants AS, Young DI, Young M, Kell DB. A bacterial cytokine. Proc Natl Acad Sci USA. 1998;95:8916–21.
Lankford CE, Walker JR, Reeves JB, Nabbut NH, Byers BR, Jones RJ. Inoculum-dependent division lag of Bacillus cultures and its relation to an endogenous factor(s) (“schizokinen”). J Bacteriol. 1966;91:1070–9.
Halmann M, Benedict M, Mager J. Nutritional Requirements of Pasteurella tularensis for Growth from Small Inocula. J Gen Microbiol. 1967;49:451–60.
Jannasch HW. Bacterial growth at low population densities. Nat. 1962;196:496–7.
Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–25.
Schincaglia GP, Hong BY, Rosania A, Barasz J, Thompson A, Sobue T, et al. Clinical, immune, and microbiome traits of gingivitis and peri-implant mucositis. J Dent Res. 2017;96:47–55.
Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–85.
Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8:471–80.
Loe H, Theilade E, Jensen SB. Experimental gingivitis in man. J Periodontol. 1965;36:177–87.
Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30–44.
Kuboniwa M, Houser JR, Hendrickson EL, Wang Q, Alghamdi SA, Sakanaka A, et al. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat Microbiol. 2017;2:1493–9.
Zhou P, Li X, Huang IH, Qi F. Veillonella catalase protects the growth of Fusobacterium nucleatum in microaerophilic and Streptococcus gordonii-resident environments. Appl Environ Microbiol. 2017;83:19.
Stacy A, Fleming D, Lamont RJ, Rumbaugh KP, Whiteley M. A commensal bacterium promotes virulence of an opportunistic pathogen via cross-respiration. mBio. 2016;7:3.
Lyons SR, Griffen AL, Leys EJ. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J Clin Microbiol. 2000;38:2362–5.
Hong BY, Furtado Araujo MV, Strausbaugh LD, Terzi E, Ioannidou E, Diaz PI. Microbiome profiles in periodontitis in relation to host and disease characteristics. PloS ONE. 2015;10:e0127077.
Tanner AC, Kent R Jr, Kanasi E, Lu SC, Paster BJ, Sonis ST, et al. Clinical characteristics and microbiota of progressing slight chronic periodontitis in adults. J Clin Periodontol. 2007;34:917–30.
Yost S, Duran-Pinedo AE, Teles R, Krishnan K, Frias-Lopez J. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 2015;7:27.
Maekawa T, Krauss JL, Abe T, Jotwani R, Triantafilou M, Triantafilou K, et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe. 2014;15:768–78.
Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506.
Lamell CW, Griffen AL, McClellan DL, Leys EJ. Acquisition and colonization stability of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in children. J Clin Microbiol. 2000;38:1196–9.
Teles FR, Teles RP, Sachdeo A, Uzel NG, Song XQ, Torresyap G, et al. Comparison of microbial changes in early redeveloping biofilms on natural teeth and dentures. J Periodontol. 2012;83:1139–48.
Naginyte M, Do T, Meade J, Devine DA, Marsh PD. Enrichment of periodontal pathogens from the biofilms of healthy adults. Sci Rep. 2019;9:5491.
Davey ME. Techniques for the growth of Porphyromonas gingivalis biofilms. Periodontol 2000. 2006;42:27–35.
James CE, Hasegawa Y, Park Y, Yeung V, Tribble GD, Kuboniwa M, et al. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infec Immun. 2006;74:3834–44.
Bizhang M, Ellerbrock B, Preza D, Raab W, Singh P, Beikler T, et al. Detection of nine microorganisms from the initial carious root lesions using a TaqMan-based real-time PCR. Oral Dis. 2011;17:642–52.
Byrne SJ, Dashper SG, Darby IB, Adams GG, Hoffmann B, Reynolds EC. Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol Immunol. 2009;24:469–77.
Huang S, Li R, Zeng X, He T, Zhao H, Chang A, et al. Predictive modeling of gingivitis severity and susceptibility via oral microbiota. ISME J. 2014;8:1768–80.
Camelo-Castillo A, Novoa L, Balsa-Castro C, Blanco J, Mira A, Tomas I. Relationship between periodontitis-associated subgingival microbiota and clinical inflammation by 16S pyrosequencing. J Clin Periodontol. 2015;42:1074–82.
Kirst ME, Li EC, Alfant B, Chi YY, Walker C, Magnusson I, et al. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl Environ Microbiol. 2015;81:783–93.
Kistler JO, Booth V, Bradshaw DJ, Wade WG. Bacterial community development in experimental gingivitis. PloS ONE. 2013;8:e71227.
The-Human-Microbiome-Project-Consortium. Structure, function and diversity of the healthy human microbiome. Nat. 2012;486:207–14.
Ganesan SM, Joshi V, Fellows M, Dabdoub SM, Nagaraja HN, O’Donnell B, et al. A tale of two risks: smoking, diabetes and the subgingival microbiome. ISME J. 2017;11:2075–89.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41.
Abe T, Hajishengallis G. Optimization of the ligature-induced periodontitis model in mice. J Immunol Methods. 2013;394:49–54.
Price RR, Viscount HB, Stanley MC, Leung KP. Targeted profiling of oral bacteria in human saliva and in vitro biofilms with quantitative real-time PCR. Biofouling 2007;23:203–13.
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60.
Albuquerque P, Nicola AM, Nieves E, Paes HC, Williamson PR, Silva-Pereira I, et al. Quorum sensing-mediated, cell density-dependent regulation of growth and virulence in Cryptococcus neoformans. mBio 2013;5:e00986–13.
Chen H, Fujita M, Feng Q, Clardy J, Fink GR. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc Natl Acad Sci USA. 2004;101:5048–52.
Yoshida M, Kashiwagi K, Shigemasa A, Taniguchi S, Yamamoto K, Makinoshima H, et al. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J Biol Chem. 2004;279:46008–13.
Dutzan N, Abusleme L, Bridgeman H, Greenwell-Wild T, Zangerle-Murray T, Fife ME, et al. On-going mechanical damage from mastication drives homeostatic Th17 cell responses at the oral barrier. Immunity 2017;46:133–47.
Kassebaum NJ, Bernabe E, Dahiya M, Bhandari B, Murray CJ, Marcenes W. Global burden of severe periodontitis in 1990-2010: a systematic review and meta-regression. J Dent Res. 2014;93:1045–53.
Zhou P, Li X, Qi F. Identification and characterization of a haem biosynthesis locus in Veillonella. Microbiol. 2016;162:1735–43.
Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16:745–59.
Baker PJ, Dixon M, Roopenian DC. Genetic control of susceptibility to Porphyromonas gingivalis-induced alveolar bone loss in mice. Infec Immun. 2000;68:5864–8.
Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, et al. Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. 2019;5:eaau3333.
Acknowledgements
This work was supported by grants R21DE023967 (PID) and R01DE015254 (GH) and by the Intramural Program (NMM) of The National Institute of Dental and Craniofacial Research, National Institutes of Health. We also acknowledge the support of the Chilean National Fund for Scientific and Technologic Development (FONDECYT) grant 11180505 (LA).
Author information
Authors and Affiliations
Contributions
PID, NMM, PDM and GH contributed to study design and supervised research. AH, HW and AM performed experiments. PID, AH, LA and BYH analyzed data. PID and AH drafted the manuscript. All authors read, critically revised and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Hoare, A., Wang, H., Meethil, A. et al. A cross-species interaction with a symbiotic commensal enables cell-density-dependent growth and in vivo virulence of an oral pathogen. ISME J 15, 1490–1504 (2021). https://doi.org/10.1038/s41396-020-00865-y
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41396-020-00865-y
This article is cited by
-
Causal association between oral microbiota and oral cancer: a mendelian randomization study
Scientific Reports (2025)
-
Bacterial translocation signatures and subgingival microbiome in individuals with periodontitis
Clinical Oral Investigations (2025)
-
Elevated fecal calprotectin is associated with gut microbial dysbiosis, altered serum markers and clinical outcomes in older individuals
Scientific Reports (2024)
-
Oral Health in Patients with History of Head and Neck Cancer: Complexity and Benefits of a Targeted Oral Healthcare Pathway
Current Oncology Reports (2024)
-
Phages are unrecognized players in the ecology of the oral pathogen Porphyromonas gingivalis
Microbiome (2023)