Abstract
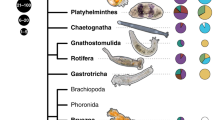
Microbiome assemblages of plants and animals often show a degree of correlation with host phylogeny; an eco-evolutionary pattern known as phylosymbiosis. Using 16S rRNA gene sequencing to profile the microbiome, paired with COI, 18S rRNA and ITS1 host phylogenies, phylosymbiosis was investigated in four groups of coral reef invertebrates (scleractinian corals, octocorals, sponges and ascidians). We tested three commonly used metrics to evaluate the extent of phylosymbiosis: (a) intraspecific versus interspecific microbiome variation, (b) topological comparisons between host phylogeny and hierarchical clustering (dendrogram) of host-associated microbial communities, and (c) correlation of host phylogenetic distance with microbial community dissimilarity. In all instances, intraspecific variation in microbiome composition was significantly lower than interspecific variation. Similarly, topological congruency between host phylogeny and the associated microbial dendrogram was more significant than would be expected by chance across all groups, except when using unweighted UniFrac distance (compared with weighted UniFrac and Bray–Curtis dissimilarity). Interestingly, all but the ascidians showed a significant positive correlation between host phylogenetic distance and associated microbial dissimilarity. Our findings provide new perspectives on the diverse nature of marine phylosymbioses and the complex roles of the microbiome in the evolution of marine invertebrates.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
All microbial data have been made available at the NCBI Sequence Read Archive under the BioProject accession number PRJNA577361 and host sequence data are available at the CNGB Sequence Archive under the accession numbers N_000000252.1–N_000000348.1.
Code availability
Code used for the analysis is available at https://github.com/paobrien.
References
Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 2016;14:1–29.
Brucker RM, Bordenstein SR. The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus nasonia. Science. 2013;341:667–9.
Lim SJ, Bordenstein SR. An introduction to phylosymbiosis. Proc Biol Sci. 2020;287:20192900.
Brucker RM, Bordenstein SR. The roles of host evolutionary relationships (genus: Nasonia) and development in sturcturing microbial communities. Evolution. 2011;66:349–62.
Ochman H, Worobey M, Kuo CH, Ndjango JBN, Peeters M, Hahn BH, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:3–10.
Kohl KD, Dearing MD, Bordenstein SR. Microbial communities exhibit host species distinguishability and phylosymbiosis along the length of the gastrointestinal tract. Mol Ecol. 2018;27:1874–83.
Ross AA, Müller KM, Weese JS, Neufeld JD. Comprehensive skin microbiome analysis reveals the uniqueness of human skin and evidence for phylosymbiosis within the class Mammalia. Proc Natl Acad Sci. 2018;115:1–10.
Pollock FJ, McMinds R, Smith S, Bourne DG, Willis BL, Medina M, et al. Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat Commun. 2018;9:1–13.
Yeoh YK, Dennis PG, Paungfoo-Lonhienne C, Weber L, Brackin R, Ragan MA, et al. Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat Commun. 2017;8:1–9.
Leigh BA, Bordenstein SR, Brooks AW, Mikaelyan A, Bordenstein SR. Finer-scale phylosymbiosis: insights from insect viromes. mSystems. 2018;3:e00131–18.
van Opstal EJ, Bordenstein SR. Phylosymbiosis impacts adaptive traits in Nasonia wasps. MBio. 2019;10:1–11.
Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TCG. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci. 2013;110:E3730–8.
Douglas AE, Werren JH. Holes in the hologenome: why host-microbe symbioses are not holobionts. MBio. 2016;7:e02099–15.
Mazel F, Davis KM, Loudon A, Kwong WK, Groussin M, Parfrey LW. Is host filtering the main driver of phylosymbiosis across the tree of life? mSystems. 2018;3:e00097–18.
Groussin M, Mazel F, Sanders JG, Smillie CS, Lavergne S, Thuiller W, et al. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat Commun. 2017;8:1–12.
Hird SM, Sánchez C, Carstens BC, Brumfield RT. Comparative gut microbiota of 59 Neotropical bird species. Front Microbiol. 2015;6:1–16.
Song SJ, Sanders JG, Delsuc F, Metcalf J, Amato K, Taylor MW, et al. Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. MBio. 2020;11:1–14.
Thomas T, Moitinho-Silva L, Lurgi M, Björk JR, Easson C, Astudillo-García C, et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun. 2016;7:1–12.
Glasl B, Smith CE, Bourne DG, Webster NS. Disentangling the effect of host-genotype and environment on the microbiome of the coral Acropora tenuis. PeerJ. 2019;7:1–18.
Amato KR, G. Sanders J, Song SJ, Nute M, Metcalf JL, Thompson LR, et al. Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 2019;13:576–87.
Amato KR, Mallott EK, McDonald D, Dominy NJ, Goldberg T, Lambert JE, et al. Convergence of human and Old World monkey gut microbiomes demonstrates the importance of human ecology over phylogeny. Genome Biol. 2019;20:1–12.
Sharp KH, Distel D, Paul VJ. Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 2012;6:790–801.
Apprill A, Marlow HQ, Martindale MQ, Rappé MS. The onset of microbial associations in the coral Pocillopora meandrina. ISME J. 2009;3:685–99.
Webster NS, Taylor MW, Behnam F, Lücker S, Rattei T, Whalan S, et al. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol. 2010;12:2070–82.
O’Brien PA, Webster NS, Miller DJ, Bourne DG. Host-microbe coevolution: applying evidence from model systems to complex marine invertebrate holobionts. MBio. 2019;10:1–14.
Morrow KM, Bourne DG, Humphrey C, Botté ES, Laffy P, Zaneveld J, et al. Natural volcanic CO2seeps reveal future trajectories for host-microbial associations in corals and sponges. ISME J. 2014;9:1–15.
Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun. 2017;8:1–8.
Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14.
Apprill A, Mcnally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Micro Ecol. 2015;75:129–37.
Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Piredda R, Tomasino MP, D’Erchia AM, Manzari C, Pesole G, Montresor M, et al. Diversity and temporal patterns of planktonic protist assemblages at a Mediterranean Long Term Ecological Research site. FEMS Microbiol Ecol. 2017;93:fiw200.
Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interact sequence choice and visualization. Brief Bioinform. 2019;20:1160–6.
Yang C, Tan S, Meng G, Bourne DG, O’Brien PA, Xu J, et al. Access COI barcode efficiently using high throughput Single-End 400 bp sequencing. bioRxiv. 2018:498618.
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3.
Janssen S, Mcdonald D, Gonzalez A, Navas-Molina JA, Jiang L, Xu ZZ, et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems. 2018;3:1–14.
Team RC. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2018. https://www.r-project.org/.
McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:1–11.
Oksanen J, Blanchet G, Friendly M, Kindt R, Legendre P. McGlinn D, et al. vegan: community ecology package. R package version. 2019;2:5–4.
Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. http://ggplot2.org.
Yu G, Smith D, Zhu H, Guan Y, Tsan-Yuk Lam T. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36.
Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2018;35:526–8.
Schliep K. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–3.
Wickham H, François R, Henry L, Müller K. dplyr: a grammar of data manipulation. R package version 0.8.0.1. 2019. https://CRAN.R-project.org/package=dplyr.
McKnight DT, Huerlimann R, Bower DS, Schwarzkopf L, Alford RA, Zenger KR. Methods for normalizing microbiome data: an ecological perspective. Methods Ecol Evol. 2019;10:389–400.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–3.
Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772.
Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42.
Pöppe J, Sutcliffe P, Hooper JNA, Wörheide G, Erpenbeck D. CO I barcoding reveals new clades and radiation patterns of indo-pacific sponges of the family irciniidae (Demospongiae: Dictyoceratida). PLoS One. 2010;5:1–6.
Erwin PM, Coma R, Paula L, Serrano E, Ribes M, Mar P. Stable symbionts across the HMA-LMA dichotomy: low seasonal and interannual variation in sponge-associated bacteria from taxonomically. FEMS Microbiol Ecol. 2015;91:1–11.
Glasl B, Smith CE, Bourne DG, Webster NS. Exploring the diversity-stability paradigm using sponge microbial communities. Sci Rep. 2018;8:1–9.
Luter HM, Whalan S, Webster NS. Thermal and sedimentation stress are unlikely causes of brown spot syndrome in the coral reef sponge, Ianthella basta. PLoS One. 2012;7:e39779.
Easson CG, Thacker RW. Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front Microbiol. 2014;5:1–11.
Meyer JL, Paul VJ, Teplitski M. Community shifts in the surface microbiomes of the coral porites astreoides with unusual lesions. PLoS One. 2014;9:e100316.
Bourne D, Iida Y, Uthicke S, Smith-keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008;2:350–63.
Pollock FJ, Lamb JB, Water JAJMVan De, Smith HA, Schaffelke B, Willis BL. et al. Reduced diversity and stability of coral-associated bacterial communities and suppressed immune function precedes disease onset in corals. R Soc Open Sci. 2019;6:190355
Glasl B, Herndl GJ, Frade PR. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 2016;10:2280–92.
Work TM, Aeby GS. Microbial aggregates within tissues infect a diversity of corals throughout the Indo-Pacific. Mar Ecol Prog Ser. 2014;500:1–9.
Wada N, Ishimochi M, Matsui T, Pollock FJ, Tang S, Ainsworth TD, et al. Characterization of coral-associated microbial aggregates (CAMAs) within tissues of the coral Acropora hyacinthus. Sci Rep. 2019;9:1–13.
Sunagawa S, Woodley CM, Medina M. Threatened corals provide underexplored microbial habitats. PLoS One. 2010;5:1–7.
Van De Water JAJM, Allemand D, Ferrier-Pagès C. Host-microbe interactions in octocoral holobionts—recent advances and perspectives. Microbiome. 2018;6:1–28.
McFadden CS, Sánchez JA, France SC. Molecular phylogenetic insights into the evolution of octocorallia: a review. Integr Comp Biol. 2010;50:389–410.
Quattrini AM, Wu T, Soong K, Jeng M, Benayahu Y, Mcfadden CS. A next generation approach to species delimitation reveals the role of hybridization in a cryptic species complex of corals. BMC Evol Biol. 2019;19:1–19.
Li J, Chen Q, Long LJ, Dong JD, Yang J, Zhang S. Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci Rep. 2014;4:1–8.
Hernandez-Agreda A, Leggat W, Ainsworth TD, Hernandez-Agreda A. A comparative analysisof microbial DNA preparation methods for use with massive and branching coralgrowth forms. Front Microbiol. 2018;9:1–14.
Acknowledgements
The authors would like to thank Katharina Fabricius, Georgina Torras and Bettina Glasl for their assistance in the field. We also thank Orpheus Island Research Station and the Molecular Ecology and Evolution Laboratory for facilitating field and laboratory work. We thank Zhenyu Peng, Guohai Hu, Bo Wang, Xudan Li, Wei Zhou, Sha Liao and Junqiang Xu for providing SE400 sequencing. Guohai Hu and Bo Wang are affiliated with Guangdong Provincial Key Laboratory of Genome Read and Write (No. 2017B030301011). We also thank Long Zhou and Qiye Li for coordinating the project. This work was funded by the Beijing Genome Institute, Earthwatch Institute and Mitsubishi Corporation. PAO is supported by an AIMS@JCU postgraduate research scholarship.
Author information
Authors and Affiliations
Contributions
PAO, DGB, NSW, DJM and GZ conceived and developed the study. PAO, DGB, NSW, PRF and HAS contributed to field work. PAO, ST, CY and HAS contributed to molecular lab work. PAO analysed the microbial data and generated figures and HAS finalised the figures. PAO, ST, CY and NA analysed the phylogenetic data. PAO drafted the paper and all authors revised the paper and approved the final version.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
O’Brien, P.A., Tan, S., Yang, C. et al. Diverse coral reef invertebrates exhibit patterns of phylosymbiosis. ISME J 14, 2211–2222 (2020). https://doi.org/10.1038/s41396-020-0671-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-020-0671-x
This article is cited by
-
Protein signatures predict coral resilience and survival to thermal bleaching events
Communications Earth & Environment (2025)
-
High diversity of crustose coralline algae microbiomes across species and islands, and implications for coral recruits
Environmental Microbiome (2024)
-
Insights into the occurrence of phylosymbiosis and co-phylogeny in the holobionts of octocorals from the Mediterranean Sea and Red Sea
Animal Microbiome (2024)
-
Environmental microbial reservoir influences the bacterial communities associated with Hydra oligactis
Scientific Reports (2024)
-
Decoupling of strain- and intrastrain-level interactions of microbiomes in a sponge holobiont
Nature Communications (2024)