Abstract
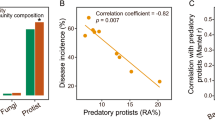
Plant health is strongly impacted by beneficial and pathogenic plant microbes, which are themselves structured by resource inputs. Organic fertilizer inputs may thus offer a means of steering soil-borne microbes, thereby affecting plant health. Concurrently, soil microbes are subject to top-down control by predators, particularly protists. However, little is known regarding the impact of microbiome predators on plant health-influencing microbes and the interactive links to plant health. Here, we aimed to decipher the importance of predator-prey interactions in influencing plant health. To achieve this goal, we investigated soil and root-associated microbiomes (bacteria, fungi and protists) over nine years of banana planting under conventional and organic fertilization regimes differing in Fusarium wilt disease incidence. We found that the reduced disease incidence and improved yield associated with organic fertilization could be best explained by higher abundances of protists and pathogen-suppressive bacteria (e.g. Bacillus spp.). The pathogen-suppressive actions of predatory protists and Bacillus spp. were mainly determined by their interactions that increased the relative abundance of secondary metabolite Q genes (e.g. nonribosomal peptide synthetase gene) within the microbiome. In a subsequent microcosm assay, we tested the interactions between predatory protists and pathogen-suppressive Bacillus spp. that showed strong improvements in plant defense. Our study shows how protistan predators stimulate disease-suppressive bacteria in the plant microbiome, ultimately enhancing plant health and yield. Thus, we suggest a new biological model useful for improving sustainable agricultural practices that is based on complex interactions between different domains of life.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
All raw 16 S rRNA, ITS and 18 S rRNA gene sequences are available at the NCBI Sequence Read Archive (SRA) under the accession number BioProject PRJNA737165. The raw data of metagenomics-derived gene catalogues are publicly available under the accession number BioProject PRJNA736854.
References
Amundson R, Berhe AA, Hopmans JW, Olson C, Sztein AE, Sparks DL. Soil and human security in the 21st century. Science. 2015;348:1261071.
Borrelli P, Robinson DA, Fleischer LR, Lugato E, Ballabio C, Alewell C, et al. An assessment of the global impact of 21st century land use change on soil erosion. Nat Commun. 2017;8:2013.
Carvalho FP. Pesticides, environment, and food safety. Food Energy Secur. 2017;6:48–60.
Santos VB, Araújo ASF, Leite LFC, Nunes LAPL, Melo WJ. Soil microbial biomass and organic matter fractions during transition from conventional to organic farming systems. Geoderma. 2012;170:227–31.
Tilman D, Fargione J, Wolff B, D’Antonio C, Dobson A, Howarth R, et al. Forecasting agriculturally driven global environmental change. Science. 2001;292:281–4.
Tu C, Louws FJ, Creamer NG, Paul Mueller J, Brownie C, Fager K, et al. Responses of soil microbial biomass and N availability to transition strategies from conventional to organic farming systems. Agric Ecosyst Environ. 2006;113:206–15.
Blundell R, Schmidt JE, Igwe A, Cheung AL, Vannette RL, Gaudin ACM, et al. Organic management promotes natural pest control through altered plant resistance to insects. Nat Plants. 2020;6:483–91.
Verbruggen E, Röling WFM, Gamper HA, Kowalchuk GA, Verhoef HA, van der Heijden MGA. Positive effects of organic farming on below-ground mutualists: large-scale comparison of mycorrhizal fungal communities in agricultural soils. N. Phytol. 2010;186:968–79.
Lupatini M, Korthals GW, de Hollander M, Janssens TKS, Kuramae EE. Soil microbiome is more heterogeneous in organic than in conventional farming system. Front Microbiol. 2017;7:2064.
Cheng H, Zhang D, Ren L, Song Z, Li Q, Wu J, et al. Bio-activation of soil with beneficial microbes after soil fumigation reduces soil-borne pathogens and increases tomato yield. Environ Pollut. 2021;283:117160.
Shahi DK, Kachhap S, Kumar A, Agarwal BK. Organic agriculture for plant disease management. In: Singh KP, Jahagirdar S, Sarma BK. (eds). Emerging Trends in Plant Pathology. 2021. Springer, Singapore, pp 643–62.
Francioli D, Schulz E, Lentendu G, Wubet T, Buscot F, Reitz T. Mineral vs organic amendments: microbial community structure, activity and abundance of agriculturally relevant microbes are driven by long-term fertilization strategies. Front Microbiol. 2016;7:1446.
Sanchez-Barrios A, Sahib MR, DeBolt S. “I’ve got the magic in me”: the microbiome of conventional vs organic production systems. In: Singh DP, Singh HB, Prabha R. (eds). Plant-Microbe Interactions in Agro-Ecological Perspectives: Volume 1: Fundamental Mechanisms, Methods and Functions. 2017. Springer, Singapore, pp 85–95.
Chowdhury SP, Babin D, Sandmann M, Jacquiod S, Sommermann L, Sørensen SJ, et al. Effect of long-term organic and mineral fertilization strategies on rhizosphere microbiota assemblage and performance of lettuce. Environ Microbiol. 2019;21:2426–39.
Weller DM. Pseudomonas biocontrol agents of soilborne pathogens: Looking back over 30 years. Phytopathology 2007;97:250–6.
Tao C, Li R, Xiong W, Shen Z, Liu S, Wang B, et al. Bio-organic fertilizers stimulate indigenous soil Pseudomonas populations to enhance plant disease suppression. Microbiome 2020;8:137.
Mazurier S, Corberand T, Lemanceau P, Raaijmakers JM. Phenazine antibiotics produced by fluorescent Pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J. 2009;3:977–91.
Yuan J, Zhao M, Li R, Huang Q, Rensing C, Shen Q. Lipopeptides produced by B. amyloliquefaciens NJN-6 altered the soil fungal community and non-ribosomal peptides genes harboring microbial community. Appl Soil Ecol. 2017;117–8:96–105.
Kiesewalter HT, Lozano-Andrade CN, Strube ML, Kovács ÁT. Secondary metabolites of Bacillus subtilis impact the assembly of soil-derived semisynthetic bacterial communities. Beilstein J Org Chem. 2020;16:2983–98.
Banerjee S, Schlaeppi K, van der Heijden MGA. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol. 2018;16:567–76.
Zhang Z, Han X, Yan J, Zou W, Wang E, Lu X, et al. Keystone microbiomes revealed by 14 years of field restoration of the degraded agricultural soil under distinct vegetation scenarios. Front Microbiol. 2020;11:1915.
Shang X, Cai X, Zhou Y, Han X, Zhang C-S, Ilyas N, et al. Pseudomonas inoculation stimulates endophytic Azospira population and induces systemic resistance to bacterial wilt. Front Plant Sci. 2021;12:1964.
Tyc O, Song C, Dickschat JS, Vos M, Garbeva P. The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends Microbiol. 2017;25:280–92.
Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285–93.
Berg G, Mahnert A, Moissl-Eichinger C. Beneficial effects of plant-associated microbes on indoor microbiomes and human health? Front Microbiol. 2014;5:15.
Straight PD, Willey JM, Kolter R. Interactions between Streptomyces coelicolor and Bacillus subtilis: Role of surfactants in raising aerial structures. J Bacteriol. 2006;188:4918–25.
González O, Ortíz-Castro R, Díaz-Pérez C, Díaz-Pérez AL, Magaña-Dueñas V, López-Bucio J, et al. Non-ribosomal peptide synthases from Pseudomonas aeruginosa play a role in cyclodipeptide biosynthesis, quorum-sensing regulation, and root development in a plant host. Micro Ecol. 2017;73:616–29.
Zhao M, Yuan J, Zhang R, Dong M, Deng X, Zhu C, et al. Microflora that harbor the NRPS gene are responsible for Fusarium wilt disease-suppressive soil. Appl Soil Ecol. 2018;132:83–90.
Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front Microbiol. 2019;10:302.
Tambadou F, Lanneluc I, Sablé S, Klein GL, Doghri I, Sopéna V, et al. Novel nonribosomal peptide synthetase (NRPS) genes sequenced from intertidal mudflat bacteria. FEMS Microbiol Lett. 2014;357:123–30.
Prieto C. Characterization of nonribosomal peptide synthetases with NRPSsp. In: Evans BS. (ed). Nonribosomal Peptide and Polyketide Biosynthesis: Methods and Protocols. 2016. Springer, New York, NY, pp 273–8.
Yuan J, Ruan Y, Wang B, Zhang J, Waseem R, Huang Q, et al. Plant growth-promoting rhizobacteria strain Bacillus amyloliquefaciens NJN-6-enriched bio-organic fertilizer suppressed Fusarium wilt and promoted the growth of banana plants. J Agric Food Chem. 2013;61:3774–80.
Yuan J, Li B, Zhang N, Waseem R, Shen Q, Huang Q. Production of bacillomycin- and macrolactin-type antibiotics by Bacillus amyloliquefaciens NJN-6 for suppressing soilborne plant pathogens. J Agric Food Chem. 2012;60:2976–81.
Xiong W, Song Y, Yang K, Gu Y, Wei Z, Kowalchuk GA, et al. Rhizosphere protists are key determinants of plant health. Microbiome. 2020;8:27.
Thakur MP, Geisen S. Trophic regulations of the soil microbiome. Trends Microbiol. 2019;27:771–80.
Müller MS, Scheu S, Jousset A. Protozoa drive the dynamics of culturable biocontrol bacterial communities. PLOS ONE. 2013;8:e66200.
Geisen S, Mitchell EAD, Adl S, Bonkowski M, Dunthorn M, Ekelund F, et al. Soil protists: A fertile frontier in soil biology research. FEMS Microbiol Rev. 2018;42:293–323.
Gao Z, Karlsson I, Geisen S, Kowalchuk G, Jousset A. Protists: puppet masters of the rhizosphere microbiome. Trends Plant Sci. 2019;24:165–76.
Jousset A, Lara E, Wall LG, Valverde C. Secondary metabolites help biocontrol strain Pseudomonas fluorescens CHA0 to escape protozoan grazing. Appl Environ Microbiol. 2006;72:7083–90.
Liu H, Xiong W, Zhang R, Hang X, Wang D, Li R, et al. Continuous application of different organic additives can suppress tomato disease by inducing the healthy rhizospheric microbiota through alterations to the bulk soil microflora. Plant Soil. 2018;423:229–40.
Chen D, Wang X, Zhang W, Zhou Z, Ding C, Liao Y, et al. Persistent organic fertilization reinforces soil-borne disease suppressiveness of rhizosphere bacterial community. Plant Soil. 2020;452:313–28.
Müller JP, Hauzy C, Hulot FD. Ingredients for protist coexistence: Competition, endosymbiosis and a pinch of biochemical interactions. J Anim Ecol. 2012;81:222–32.
Guo S, Xiong W, Hang X, Gao Z, Jiao Z, Liu H, et al. Protists as main indicators and determinants of plant performance. Microbiome. 2021;9:64.
Ren F, Sun N, Xu M, Zhang X, Wu L, Xu M. Changes in soil microbial biomass with manure application in cropping systems: a meta-analysis. Soil Tillage Res. 2019;194:104291.
Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–86.
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil. 2009;321:341–61.
Compant S, Cambon MC, Vacher C, Mitter B, Samad A, Sessitsch A. The plant endosphere world – bacterial life within plants. Environ Microbiol. 2021;23:1812–29.
Oliverio AM, Geisen S, Delgado-Baquerizo M, Maestre FT, Turner BL, Fierer N. The global-scale distributions of soil protists and their contributions to belowground systems. Sci Adv. 2020;6:eaax8787.
Dumack K, Fiore-Donno AM, Bass D, Bonkowski M. Making sense of environmental sequencing data: ecologically important functional traits of the protistan groups Cercozoa and Endomyxa (Rhizaria). Mol Ecol Resour. 2020;20:398–403.
Romdhane S, Spor A, Banerjee S, Breuil M-C, Bru D, Chabbi A, et al. Land-use intensification differentially affects bacterial, fungal and protist communities and decreases microbiome network complexity. Environ Microbiome. 2022;17:1.
Jousset A, Rochat L, Péchy-Tarr M, Keel C, Scheu S, Bonkowski M. Predators promote defence of rhizosphere bacterial populations by selective feeding on non-toxic cheaters. ISME J. 2009;3:666–74.
Yu GY, Sinclair JB, Hartman GL, Bertagnolli BL. Production of iturin A by Bacillus amyloliquefaciens suppressing Rhizoctonia solani. Soil Biol Biochem. 2002;34:955–63.
Romero D, de Vicente A, Rakotoaly RH, Dufour SE, Veening J-W, Arrebola E, et al. The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol Plant Microbe Interact. 2007;20:430–40.
Xu Z, Mandic-Mulec I, Zhang H, Liu Y, Sun X, Feng H, et al. Antibiotic bacillomycin D affects iron acquisition and biofilm formation in Bacillus velezensis through a Btr-mediated FeuABC-dependent pathway. Cell Rep. 2019;29:1192–1202.e5.
Huang J, Wei Z, Tan S, Mei X, Shen Q, Xu Y. Suppression of bacterial wilt of tomato by bioorganic fertilizer made from the antibacterial compound producing strain Bacillus amyloliquefaciens HR62. J Agric Food Chem. 2014;62:10708–16.
Wang B, Shen Z, Zhang F, Raza W, Yuan J, Huang R, et al. Bacillus amyloliquefaciens strain W19 can promote growth and yield and suppress Fusarium wilt in banana under greenhouse and field conditions. Pedosphere. 2016;26:733–44.
Shen Z, Ruan Y, Chao X, Zhang J, Li R, Shen Q. Rhizosphere microbial community manipulated by 2 years of consecutive biofertilizer application associated with banana Fusarium wilt disease suppression. Biol Fertil Soils. 2015;51:553–62.
Jeger MJ, Eden-Green S, Thresh JM, Johanson A, Waller JM, Brown AE. Banana diseases. In: Gowen S. (ed). Bananas and Plantains. 1995. Springer Netherlands, Dordrecht, pp 317–81.
Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci. 2015;112:E911–20.
Fierer N, Jackson JA, Vilgalys R, Jackson RB. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol. 2005;71:4117–20.
Jiménez-Fernández D, Montes-Borrego M, Navas-Cortés JA, Jiménez-Díaz RM, Landa BB. Identification and quantification of Fusarium oxysporum in planta and soil by means of an improved specific and quantitative PCR assay. Appl Soil Ecol. 2010;46:372–82.
Mori K, Iriye R, Hirata M, Takamizawa K. Quantification of Bacillus species in a wastewater treatment system by the molecular analyses. Biotechnol Bioprocess Eng. 2004;9:482–9.
Ayuso-Sacido A, Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Micro Ecol. 2005;49:10–24.
Fu L, Penton CR, Ruan Y, Shen Z, Xue C, Li R, et al. Inducing the rhizosphere microbiome by biofertilizer application to suppress banana Fusarium wilt disease. Soil Biol Biochem. 2017;104:39–48.
Claesson MJ, O’Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLOS ONE. 2009;4:e6669.
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds). PCR Protocols. 1990. Academic Press, San Diego, pp 315–22.
Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–8.
Bass D, Silberman JD, Brown MW, Pearce RA, Tice AK, Jousset A, et al. Coprophilic amoebae and flagellates, including Guttulinopsis, Rosculus and Helkesimastix, characterise a divergent and diverse rhizarian radiation and contribute to a large diversity of faecal-associated protists. Environ Microbiol. 2016;18:1604–19.
Geisen S, Vaulot D, Mahé F, Lara E, Vargas C de, Bass D. A user guide to environmental protistology: primers, metabarcoding, sequencing, and analyses. BioRxiv 2019;850610:1–34.
Xiong W, Jousset A, Li R, Delgado-Baquerizo M, Bahram M, Logares R, et al. A global overview of the trophic structure within microbiomes across ecosystems. Environ Int. 2021;151:106438.
Xiong W, Li R, Ren Y, Liu C, Zhao Q, Wu H, et al. Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol Biochem. 2017;107:198–207.
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200.
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7.
Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L, et al. The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013;41:D597–D604.
Xiong W, Li R, Guo S, Karlsson I, Jiao Z, Xun W, et al. Microbial amendments alter protist communities within the soil microbiome. Soil Biol Biochem. 2019;135:379–82.
Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–93.
Revelle W, Revelle MW. Package ‘psych’. Compr R Arch Netw. 2015;337:338.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300.
Bargabus RL, Zidack NK, Sherwood JE, Jacobsen BJ. Characterisation of systemic resistance in sugar beet elicited by a non-pathogenic, phyllosphere-colonizing Bacillus mycoides, biological control agent. Physiol Mol Plant Pathol. 2002;61:289–98.
Bais HP, Fall R, Vivanco JM. Biocontrol of Bacillus subtilis against infection of arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004;134:307–19.
Cazorla FM, Romero D, Pérez-García A, Lugtenberg BJJ, Vicente Ade, Bloemberg G. Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity. J Appl Microbiol. 2007;103:1950–9.
Aneja KR. Experiments in microbiology, plant pathology and biotechnology. 2007. New Age International, New Delhi.
Mela F, Fritsche K, de Boer W, van Veen JA, de Graaff LH, van den Berg M, et al. Dual transcriptional profiling of a bacterial/fungal confrontation: Collimonas fungivorans versus Aspergillus niger. ISME J. 2011;5:1494–504.
Gao Z. Soil protists: From traits to ecological functions. 2020. Utrecht University.
Anderson MJ. Permutational multivariate analysis of variance (PERMANOVA). Wiley StatsRef: Statistics Reference Online. 2017. American Cancer Society, pp 1–15.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara RB, et al. Package ‘vegan’. Community Ecol Package Version. 2013;2:1–295.
Breiman L. Random forests. Mach Learn. 2001;45:5–32.
Liaw A, Wiener M. Classification and regression by randomForest. R N. 2002;23:18–22.
Archer E. rfPermute: Estimate permutation p-values for random forest importance metrics. R Package Version 20 2016.
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60.
Oguntunde PG, Fosu M, Ajayi AE, van de Giesen N. Effects of charcoal production on maize yield, chemical properties and texture of soil. Biol Fertil Soils. 2004;39:295–9.
Mcdonald JH. Handbook of biological statistics. 2009. Baltimore: sparky house publishing, Baltimore.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (42090065, 31972509, 41867006 and 32102475), the Fundamental Research Funds for the Central Universities (KYXK202009), the China Postdoctoral Science Foundation (2021TQ0156 and 2021M691613), the 111 project (B12009), and the Priority Academic Program Development of the Jiangsu Higher Education Institutions (PAPD). Stefan Geisen was supported by an NWO-VENI grant from the Netherlands Organisation for Scientific Research (016.Veni.181.078).
Author information
Authors and Affiliations
Contributions
SG, CT, AJ, WX, ZX, ZG, RL, QS, GAK and SG developed the ideas and designed the experimental plans. SG, CT, ZW, ZS, BW, SL, RL and YR performed the experiments. SG, CT, WX, RL and SG analyzed the data. SG, CT, AJ, RL, QS, GAK and SG participated in the completion of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Guo, S., Tao, C., Jousset, A. et al. Trophic interactions between predatory protists and pathogen-suppressive bacteria impact plant health. ISME J 16, 1932–1943 (2022). https://doi.org/10.1038/s41396-022-01244-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41396-022-01244-5
This article is cited by
-
Biofertilizer induces soil disease suppression by activating pathogen suppressive protist taxa
npj Biofilms and Microbiomes (2026)
-
Harnessing the legacy effect of soil microbiome in organic manure application for better crop performance and greenhouse gas emission
Plant and Soil (2026)
-
Structural and functional properties of bacterial communities associated with rootless duckweed (Wolffia globosa) and their effect on the Wolffia growth
Environmental Microbiome (2025)
-
Dynamics of protist and bacterial communities during the nitrogen removal by ecological floating beds of Sesuvium portulacastrum
Ecological Processes (2025)
-
Soil-dwelling Naegleria enhances plant performance by stimulating beneficial bacterial functions in the rhizosphere
Nature Communications (2025)