Abstract
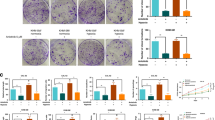
Anlotinib, a multitarget tyrosine kinase inhibitor, is effective as a third-line treatment against non-small cell lung cancer (NSCLC). However, acquired resistance occurs during its administration. To understand the molecular mechanisms of anlotinib resistance, we characterized chromatin accessibility in both the parental and anlotinib-resistant lung cancer cell line NCI-H1975 through ATAC-seq. Compared with the parental cells, we identified 2666 genomic regions with greater accessibility in anlotinib-resistant cells, in which angiogenesis-related processes and the motifs of 21 transcription factors were enriched. Among these transcription factors, TFAP2A was upregulated. TFAP2A knockdown robustly diminished tumor-induced angiogenesis and partially rescued the anti-angiogenic activity of anlotinib. Furthermore, transcriptome analysis indicated that 2280 genes were downregulated in anlotinib-resistant cells with TFAP2A knocked down, among which the PDGFR, TGF-β, and VEGFR signaling pathways were enriched. Meanwhile, we demonstrated that TFAP2A binds to accessible sites within BMP4 and HSPG2. Collectively, this study suggests that TFAP2A accelerates anlotinib resistance by promoting tumor-induced angiogenesis.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–89.
Zhang LL, Kan MY, Zhang MM, Yu SS, Xie HJ, Gu ZH, et al. Multiregion sequencing reveals the intratumor heterogeneity of driver mutations in TP53-driven non-small cell lung cancer. Int J Cancer. 2017;140:103–8.
Han B, Li K, Zhao Y, Li B, Cheng Y, Zhou J, et al. Anlotinib as a third-line therapy in patients with refractory advanced non-small-cell lung cancer: a multicentre, randomised phase II trial (ALTER0302). Br J Cancer. 2018;118:654–61.
Han B, Li K, Wang Q, Zhang L, Shi J, Wang Z, et al. Effect of anlotinib as a third-line or further treatment on overall survival of patients with advanced non-small cell lung cancer: the ALTER 0303 phase 3 randomized clinical trial. JAMA Oncol. 2018;4:1569–75.
Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357:eaal2380.
Brown R, Curry E, Magnani L, Wilhelm-Benartzi CS, Borley J. Poised epigenetic states and acquired drug resistance in cancer. Nat Rev Cancer. 2014;14:747–53.
Zhang YW, Zheng Y, Wang JZ, Lu XX, Wang Z, Chen LB, et al. Integrated analysis of DNA methylation and mRNA expression profiling reveals candidate genes associated with cisplatin resistance in non-small cell lung cancer. Epigenetics. 2014;9:896–909.
Chang XF, Monitto CL, Demokan S, Kim MS, Chang SS, Zhong XL, et al. Identification of hypermethylated genes associated with cisplatin resistance in human cancers. Cancer Res. 2010;70:2870–9.
Zeller C, Dai W, Steele NL, Siddiq A, Walley AJ, Wilhelm-Benartzi CSM, et al. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene. 2012;31:4567–76.
Tome-Garcia J, Erfani P, Nudelman G, Tsankov AM, Katsyv I, Tejero R, et al. Analysis of chromatin accessibility uncovers TEAD1 as a regulator of migration in human glioblastoma. Nat Commun. 2018;9:4020.
Denny SK, Yang D, Chuang CH, Brady JJ, Lim JS, Gruner BM, et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell. 2016;166:328–42.
Jing DH, Huang YZ, Liu XY, Sia KCS, Zhang JC, Tai XL, et al. Lymphocyte-specific chromatin accessibility pre-determines glucocorticoid resistance in acute lymphoblastic leukemia. Cancer Cell. 2018;34:906–21.
Lv ZB, Wei JL, You WX, Wang R, Shang JK, Xiong YF, et al. Disruption of the c-Myc/miR-200b-3p/PRDX2 regulatory loop enhances tumor metastasis and chemotherapeutic resistance in colorectal cancer. J Transl Med. 2017;15:257.
Ning JF, Stanciu M, Humphrey MR, Gorham J, Wakimoto H, Nishihara R, et al. Myc targeted CDK18 promotes ATR and homologous recombination to mediate PARP inhibitor resistance in glioblastoma. Nat Commun. 2019;10:2910.
Liu D, Zhang XX, Li MC, Cao CH, Wan DY, Xi BX, et al. C/EBPbeta enhances platinum resistance of ovarian cancer cells by reprogramming H3K79 methylation. Nat Commun. 2018;9:1739.
Karvonen H, Barker H, Kaleva L, Niininen W, Ungureanu D. Molecular mechanisms associated with ROR1-mediated drug resistance: crosstalk with Hippo-YAP/TAZ and BMI-1 pathways. Cells. 2019;8:812.
Huh HD, Kim DH, Jeong HS, Park HW. Regulation of TEAD transcription factors in cancer biology. Cells. 2019;8:600.
Wang S, Xiao Z, Hong Z, Jiao H, Zhu S, Zhao Y, et al. FOXF1 promotes angiogenesis and accelerates bevacizumab resistance in colorectal cancer by transcriptionally activating VEGFA. Cancer Lett. 2018;439:78–90.
Vervoort SJ, de Jong OG, Roukens MG, Frederiks CL, Vermeulen JF, Lourenco AR, et al. Global transcriptional analysis identifies a novel role for SOX4 in tumor-induced angiogenesis. Elife. 2018;7:e27706.
Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–9. https://doi.org/10.1002/0471142727.mb2129s109.
Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–62.
Wu J, Xu J, Liu B, Yao G, Wang P, Lin Z, et al. Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature. 2018;557:256–60.
de la Torre-Ubieta L, Stein JL, Won H, Opland CK, Liang D, Lu D, et al. The dynamic landscape of open chromatin during human cortical neurogenesis. Cell. 2018;172:289–304 e18.
Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–3.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Tan S, Li H, Zhang W, Shao Y, Liu Y, Guan H, et al. NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene. 2018;37:4887–900.
Zhang XL, Wu J, Wang J, Shen T, Li H, Lu J, et al. Integrative epigenomic analysis reveals unique epigenetic signatures involved in unipotency of mouse female germline stem cells. Genome Biol. 2016;17:162.
Lu J, Xu W, Qian J, Wang S, Zhang B, Zhang L, et al. Transcriptome profiling analysis reveals that CXCL2 is involved in anlotinib resistance in human lung cancer cells. BMC Med Genomics. 2019;12:38.
Lu J, Shi Q, Zhang L, Wu J, Lou Y, Qian J, et al. Integrated transcriptome analysis reveals KLK5 and L1CAM predict response to anlotinib in NSCLC at 3rd line. Front Oncol. 2019;9:886.
Gomez-Miragaya J, Moran S, Calleja-Cervantes ME, Collado-Sole A, Pare L, Gomez A, et al. The altered transcriptome and DNA methylation profiles of docetaxel resistance in breast cancer PDX models. Mol Cancer Res. 2019;17:2063–76.
Rothhammer T, Bataille F, Spruss T, Eissner G, Bosserhoff AK. Functional implication of BMP4 expression on angiogenesis in malignant melanoma. Oncogene. 2007;26:4158–70.
Dimitrova Y, Gruber AJ, Mittal N, Ghosh S, Dimitriades B, Mathow D, et al. TFAP2A is a component of the ZEB1/2 network that regulates TGFB1-induced epithelial to mesenchymal transition. Biol Direct. 2017;12:8.
Pu M, Li C, Qi X, Chen J, Wang Y, Gao L, et al. MiR-1254 suppresses HO-1 expression through seed region-dependent silencing and non-seed interaction with TFAP2A transcript to attenuate NSCLC growth. PLoS Genet. 2017;13:e1006896.
Lin B, Song X, Yang D, Bai D, Yao Y, Lu N. Anlotinib inhibits angiogenesis via suppressing the activation of VEGFR2, PDGFRbeta and FGFR1. Gene. 2018;654:77–86.
Sun YK, Niu W, Du F, Du CX, Li ST, Wang JW, et al. Safety, pharmacokinetics, and antitumor properties of anlotinib, an oral multi-target tyrosine kinase inhibitor, in patients with advanced refractory solid tumors. J Hematol Oncol. 2016;9:105.
Lu J, Zhong H, Chu T, Zhang X, Li R, Sun J, et al. Role of anlotinib-induced CCL2 decrease in anti-angiogenesis and response prediction for nonsmall cell lung cancer therapy. Eur Respir J. 2019;53:1801562.
Lu J, Zhong H, Wu J, Chu T, Zhang L, Li H, et al. Circulating DNA-based sequencing guided anlotinib therapy in non-small cell lung cancer. Adv Sci (Weinh). 2019;6:1900721.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Viallard C, Larrivee B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20:409–26.
Shi D, Xie F, Zhang Y, Tian Y, Chen W, Fu L, et al. TFAP2A regulates nasopharyngeal carcinoma growth and survival by targeting HIF-1alpha signaling pathway. Cancer Prev Res (Philos). 2014;7:266–77.
Rathinavelu A, Narasimhan M, Muthumani P. A novel regulation of VEGF expression by HIF-1a and STAT3 in HDM2 transfected prostate cancer cells. J Cell Mol Med. 2012;16:1750–7.
Suzuki Y, Montagne K, Nishihara A, Watabe T, Miyazono K. BMPs promote proliferation and migration of endothelial cells via stimulation of VEGF-A/VEGFR2 and Angiopoietin-1/Tie2 signalling. J Biochem. 2008;143:199–206.
Acknowledgements
This research was funded by the National Key Research and Development Program of China (number 2018YFC1003501), the System Biomedicine Innovation Center Program from Shanghai Jiao Tong University (number 15ZH4009), the Key Translational Medicine Program from Shanghai Jiao Tong University School of Medicine (number 15ZH1008), and the National Natural Science Foundation of China (number 81874037).
Author information
Authors and Affiliations
Contributions
LLZ, JL, and RQL performed the experiment. LLZ wrote the paper. MJH, YMZ, ST, SYW, BZ, WN, YD, and HZ helped to analyzed the data. BHH, XDZ, and WZ designed the study and performed manuscript editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Zhang, Ll., Lu, J., Liu, Rq. et al. Chromatin accessibility analysis reveals that TFAP2A promotes angiogenesis in acquired resistance to anlotinib in lung cancer cells. Acta Pharmacol Sin 41, 1357–1365 (2020). https://doi.org/10.1038/s41401-020-0421-7
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41401-020-0421-7
Keywords
This article is cited by
-
TFAP2A upregulates FAM83A to suppress ferroptosis and diminish cisplatin sensitivity in non-small cell lung cancer
Cell Division (2025)
-
TFAP2A downregulation mediates tumor-suppressive effect of miR-8072 in triple-negative breast cancer via inhibiting SNAI1 transcription
Breast Cancer Research (2024)
-
Chromatin accessibility: biological functions, molecular mechanisms and therapeutic application
Signal Transduction and Targeted Therapy (2024)
-
Crucial role of the transcription factors family activator protein 2 in cancer: current clue and views
Journal of Translational Medicine (2023)
-
A rapid and stable spontaneous reprogramming system of Spermatogonial stem cells to Pluripotent State
Cell & Bioscience (2023)