Abstract
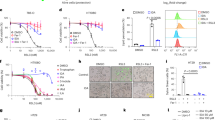
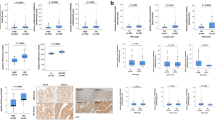
Colorectal cancer (CRC) is the third most common cancer in men and the second most common cancer in women worldwide. CRC is the second leading cause of cancer-related deaths. Although some progress in the treatment of CRC has been achieved, the molecular mechanism of CRC is still unclear. In this study, alcohol dehydrogenase 1C(ADH1C) was first identified as a target gene closely associated with the development of CRC by the comprehensive application of transcriptomics, proteomics, metabonomics and in silico analysis. The ADH1C mRNA and protein expression in CRC cell lines and tumor tissues was lower than that in normal intestinal epithelial cell lines and healthy tissues. Overexpression of ADH1C inhibited the growth, migration, invasion and colony formation of CRC cell lines and prevented the growth of xenograft tumors in nude mice. The inhibitory effects of ADH1C on CRC cells in vitro were exerted by reducing the expression of PHGDH/PSAT1 and the serine level. This inhibition could be partially reversed by adding serine to the culture medium. These results showed that ADH1C is a potential drug target in CRC.

Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Change history
25 June 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41401-024-01319-3
References
Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–502. https://doi.org/10.1016/S0140-6736(13)61649-9.
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA: a cancer journal for clinicians. 2020;70:145–64. https://doi.org/10.3322/caac.21601.
Gu MJ, Huang QC, Bao CZ, Li YJ, Li XQ, Ye D, et al. Attributable causes of colorectal cancer in China. BMC Cancer. 2018;18:38 https://doi.org/10.1186/s12885-017-3968-z.
Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669–80. https://doi.org/10.1038/s41568-021-00378-6.
Liu X, Li T, Kong D, You H, Kong F, Tang R. Prognostic implications of alcohol dehydrogenases in hepatocellular carcinoma. BMC Cancer. 2020;20:1204. https://doi.org/10.1186/s12885-020-07689-1.
Gupta MK, Behara SK, Vadde R. In silico analysis of differential gene expressions in biliary stricture and hepatic carcinoma. Gene. 2017;597:49–58. https://doi.org/10.1016/j.gene.2016.10.032.
Shen XY, Liu XP, Song CK, Wang YJ, Li S, Hu WD. Genome-wide analysis reveals alcohol dehydrogenase 1C and secreted phosphoprotein 1 for prognostic biomarkers in lung adenocarcinoma. J Cell Physiol. 2019;234:22311–20. https://doi.org/10.1002/jcp.28797.
Liang L, Liu M, Sun X, Yuan Y, Peng K, Rashid K, et al. Identification of key genes involved in tumor immune cell infiltration and cetuximab resistance in colorectal cancer. Cancer Cell Int. 2021;21:135. https://doi.org/10.1186/s12935-021-01829-8.
Ghosh S, Bankura B, Ghosh S, Saha ML, Pattanayak AK, Ghatak S, et al. Polymorphisms in ADH1B and ALDH2 genes associated with the increased risk of gastric cancer in West Bengal, India. BMC Cancer. 2017;17:782. https://doi.org/10.1186/s12885-017-3713-7.
Wang L, Zhang Y, Ding D, He X, Zhu Z. Lack of association of ADH1C genotype with breast cancer susceptibility in Caucasian population: a pooled analysis of case-control studies. Breast. 2012;21:435–9. https://doi.org/10.1016/j.breast.2012.01.007.
Wu M, Chang SC, Kampman E, Yang J, Wang XS, Gu XP, et al. Single nucleotide polymorphisms of ADH1B, ADH1C and ALDH2 genes and esophageal cancer: a population-based case-control study in China. Int J Cancer. 2013;132:1868–77. https://doi.org/10.1002/ijc.27803.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991-D5. https://doi.org/10.1093/nar/gks1193.
Lee D, Park Y, Kim S Towards multi-omics characterization of tumor heterogeneity: a comprehensive review of statistical and machine learning approaches. Brief Bioinform. 2021;22. https://doi.org/10.1093/bib/bbaa188.
Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–7.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559. https://doi.org/10.1186/1471-2105-9-559.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. https://doi.org/10.1038/nprot.2008.211.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. https://doi.org/10.1101/gr.092759.109.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8:S11. https://doi.org/10.1186/1752-0509-8-S4-S11.
Jia XQ, Zhang S, Zhu HJ, Wang W, Zhu JH, Wang XD, et al. Increased expression of PHGDH and prognostic significance in colorectal cancer. Transl Oncol. 2016;9:191–6. https://doi.org/10.1016/j.tranon.2016.03.006.
Vié N, Copois V, Bascoul-Mollevi C, Denis V, Bec N, Robert B, et al. Overexpression of phosphoserine aminotransferase PSAT1 stimulates cell growth and increases chemoresistance of colon cancer cells. Mol Cancer. 2008;7:14. https://doi.org/10.1186/1476-4598-7-14.
Deng L, Yao P, Li L, Ji F, Zhao S, Xu C, et al. p53-mediated control of aspartate-asparagine homeostasis dictates LKB1 activity and modulates cell survival. Nat Commun. 1212. 2020;11. https://doi.org/10.1038/s41467-020-15573-6.
Li S, Liu J, Zheng X, Ren L, Yang Y, Li W, et al. Tumorigenic bacteria in colorectal cancer: mechanisms and treatments. Cancer Biol Med. 2021. https://doi.org/10.20892/j.issn.2095-3941.2020.0651.
Matsuo K, Nishimura M, Komurov K, Shahzad MMK, Ali-Fehmi R, Roh JW, et al. Platelet-derived growth factor receptor alpha (PDGFRα) targeting and relevant biomarkers in ovarian carcinoma. Gynecol Oncol. 2014;132:166–75. https://doi.org/10.1016/j.ygyno.2013.10.027.
Wang YZ, Qiu SC. Prediction of key genes in ovarian cancer treated with decitabine based on network strategy. Oncol Rep. 2016;35:3548–58. https://doi.org/10.3892/or.2016.4697.
Niemira M, Collin F, Szalkowska A, Bielska A, Chwialkowska K, Reszec J, et al. Molecular signature of subtypes of non-small-cell lung cancer by large-scale transcriptional profiling: identification of key modules and genes by weighted gene co-expression network analysis (WGCNA). Cancers. 2019;12. https://doi.org/10.3390/cancers12010037.
Xiang Z, Li J, Song S, Wang J, Cai W, Hu W, et al. A positive feedback between IDO1 metabolite and COL12A1 via MAPK pathway to promote gastric cancer metastasis. J Exp Clin Cancer Res. 2019;38:314. https://doi.org/10.1186/s13046-019-1318-5.
Wang Z, Tang W, Yuan J, Qiang B, Han W, Peng X. Integrated analysis of RNA-binding proteins in glioma. Cancers. 2020;12. https://doi.org/10.3390/cancers12040892.
Yan J, Wu L, Jia C, Yu S, Lu Z, Sun Y, et al. Development of a four-gene prognostic model for pancreatic cancer based on transcriptome dysregulation. Aging. 2020;12:3747–70. https://doi.org/10.18632/aging.102844.
Lian P, Wang Q, Zhao Y, Chen C, Sun X, Li H, et al. An eight-long non-coding RNA signature as a candidate prognostic biomarker for bladder cancer. Aging. 2019;11:6930–40. https://doi.org/10.18632/aging.102225.
Teschke R. Alcoholic liver disease: alcohol metabolism, cascade of molecular mechanisms, cellular targets, and clinical aspects. Biomedicines. 2018;6. https://doi.org/10.3390/biomedicines6040106.
Rodriguez FD, Coveñas R. Biochemical mechanisms associating alcohol use disorders with cancers. Cancers. 2021;13. https://doi.org/10.3390/cancers13143548.
Crabb DW, Matsumoto M, Chang D, You M. Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proc Nutr Soc. 2004;63:49–63.
Guo B, Zhang H, Wang J, Wu R, Zhang J, Zhang Q, et al. Identification of the signature associated with mA RNA methylation regulators and mA-related genes and construction of the risk score for prognostication in early-stage lung adenocarcinoma. Front Genet. 2021;12:656114. https://doi.org/10.3389/fgene.2021.656114.
Hosios AM, Manning BD. Cancer signaling drives cancer metabolism: AKT and the Warburg effect. Cancer Res. 2021;81:4896–8. https://doi.org/10.1158/0008-5472.CAN-21-2647.
Li AM, Ye J. Reprogramming of serine, glycine and one-carbon metabolism in cancer. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165841. https://doi.org/10.1016/j.bbadis.2020.165841.
Zhang J, Pavlova NN, Thompson CB. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J. 2017;36:1302–15. https://doi.org/10.15252/embj.201696151.
DeBerardinis RJ. Serine metabolism: some tumors take the road less traveled. Cell Metab. 2011;14:285–6. https://doi.org/10.1016/j.cmet.2011.08.004.
Dolfi SC, Chan LLY, Qiu J, Tedeschi PM, Bertino JR, Hirshfield KM, et al. The metabolic demands of cancer cells are coupled to their size and protein synthesis rates. Cancer Metab. 2013;1:20. https://doi.org/10.1186/2049-3002-1-20.
Hashibe M, McKay JD, Curado MP, Oliveira JC, Koifman S, Koifman R, et al. Multiple ADH genes are associated with upper aerodigestive cancers. Nat Genet. 2008;40:707–9. https://doi.org/10.1038/ng.151.
Suo C, Yang Y, Yuan Z, Zhang T, Yang X, Qing T, et al. Alcohol intake interacts with functional genetic polymorphisms of aldehyde dehydrogenase (ALDH2) and alcohol dehydrogenase (ADH) to increase esophageal squamous cell cancer risk. J Thorac Oncol. 2019;14:712–25. https://doi.org/10.1016/j.jtho.2018.12.023.
Tan SLW, Chadha S, Liu Y, Gabasova E, Perera D, Ahmed K, et al. A class of environmental and endogenous toxins induces BRCA2 haploinsufficiency and genome instability. Cell. 2017;169. https://doi.org/10.1016/j.cell.2017.05.010.
Zhao JY, Feng KR, Wang F, Zhang JW, Cheng JF, Lin GQ, et al. A retrospective overview of PHGDH and its inhibitors for regulating cancer metabolism. Eur J Med Chem. 2021;217:113379. https://doi.org/10.1016/j.ejmech.2021.113379.
Snell K, Natsumeda Y, Eble JN, Glover JL, Weber G. Enzymic imbalance in serine metabolism in human colon carcinoma and rat sarcoma. Br J Cancer. 1988;57:87–90.
Biyik-Sit R, Kruer T, Dougherty S, Bradley JA, Wilkey DW, Merchant ML, et al. Nuclear pyruvate kinase M2 (PKM2) contributes to phosphoserine aminotransferase 1 (PSAT1)-mediated cell migration in egfr-activated lung cancer cells. Cancers. 2021;13. https://doi.org/10.3390/cancers13163938.
Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50. https://doi.org/10.1038/nature10350.
Muthusamy T, Cordes T, Handzlik MK, You L, Lim EW, Gengatharan J, et al. Serine restriction alters sphingolipid diversity to constrain tumour growth. Nature. 2020;586:790–5. https://doi.org/10.1038/s41586-020-2609-x.
Williams RT, Guarecuco R, Gates LA, Barrows D, Passarelli MC, Carey B, et al. ZBTB1 regulates asparagine synthesis and leukemia cell response to L-asparaginase. Cell Metab. 2020;31:852–61.e6. https://doi.org/10.1016/j.cmet.2020.03.008.
Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci USA. 2012;109:6904–9. https://doi.org/10.1073/pnas.1204176109.
Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–96. https://doi.org/10.1038/emboj.2010.81.
Acknowledgements
This research was funded by Beijing Natural Science Foundation (7212157), CAMS Innovation Fund for Medical Sciences (2021-1-I2M-029), National Natural Science Foundation of China (81803584, 81703536), Technology Major Projects for “Major New Drugs Innovation and Development” (2018ZX09711001-005-025, 2018ZX09711001-012), National Natural Science Foundation of China (82073311).
Author information
Authors and Affiliations
Contributions
JHW and GHD developed the hypothesis, designed the experiments, and revised the manuscript. SL conducted most experiments and wrote the main manuscript. HY, JYL and LWR accomplished the rest experiments, WL, YHY, BBG, YZZ, WQF and XJZ collected data and performed the statistical analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
The original online version of this article was revised: errors in the previously published images of Fig. 6d, e and Suppl Fig. 8c,d. These inaccuracies were due to an oversight during the preparation of the figures.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, S., Yang, H., Li, W. et al. ADH1C inhibits progression of colorectal cancer through the ADH1C/PHGDH /PSAT1/serine metabolic pathway. Acta Pharmacol Sin 43, 2709–2722 (2022). https://doi.org/10.1038/s41401-022-00894-7
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41401-022-00894-7
Keywords
This article is cited by
-
Bioinformatics analysis of ferroptosis-related hub genes and immunoinfiltration in myocardial ischemia/reperfusion following heart transplantation
BMC Cardiovascular Disorders (2025)
-
Lysosome-related molecular subtypes reveal prognostic signatures and immunotherapeutic implications in ovarian cancer
Scientific Reports (2025)
-
PSAT1 inhibits ferroptosis in osteosarcoma cells by activating the Xct/GPX4 signaling axis
Scientific Reports (2025)
-
PSAT1 promotes the progression of colorectal cancer by regulating Hippo-YAP/TAZ-ID1 axis via AMOT
Molecular and Cellular Biochemistry (2025)
-
Mechanistic study on the inhibition of triple-negative breast cancer progression by LncRNA TFAP2A-AS1 through the regulation of miR-6892/PHGDH
Inflammation Research (2025)