Abstract
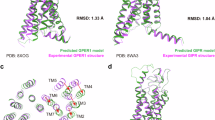
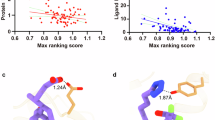
As important drug targets, G protein-coupled receptors (GPCRs) play pivotal roles in a wide range of physiological processes. Extensive efforts of structural biology have been made on the study of GPCRs. However, a large portion of GPCR structures remain unsolved due to structural instability. Recently, AlphaFold2 has been developed to predict structure models of many functionally important proteins including all members of the GPCR family. Herein we evaluated the accuracy of GPCR structure models predicted by AlphaFold2. We revealed that AlphaFold2 could capture the overall backbone features of the receptors. However, the predicted models and experimental structures were different in many aspects including the assembly of the extracellular and transmembrane domains, the shape of the ligand-binding pockets, and the conformation of the transducer-binding interfaces. These differences impeded the use of predicted structure models in the functional study and structure-based drug design of GPCRs, which required reliable high-resolution structural information.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–51.
Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–7.
Xu P, Huang S, Zhang H, Mao C, Zhou XE, Cheng X, et al. Structural insights into the lipid and ligand regulation of serotonin receptors. Nature. 2021;592:469–73.
Duan J, Xu P, Cheng X, Mao C, Croll T, He X, et al. Structures of full-length glycoprotein hormone receptor signalling complexes. Nature. 2021;598:688–92.
Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–42.
Kooistra AJ, Mordalski S, Pándy-Szekeres G, Esguerra M, Mamyrbekov A, Munk C, et al. GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Res. 2021;49:D335–43.
Thal DM, Vuckovic Z, Draper-Joyce CJ, Liang YL, Glukhova A, Christopoulos A, et al. Recent advances in the determination of G protein-coupled receptor structures. Curr Opin Struct Biol. 2018;51:28–34.
Safdari HA, Pandey S, Shukla AK, Dutta S. Illuminating GPCR signaling by Cryo-EM. Trends Cell Biol. 2018;28:591–4.
Duan J, Shen DD, Zhou XE, Bi P, Liu QF, Tan YX, et al. Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy. Nat Commun. 2020;11:4121.
Rose PW, Prlić A, Altunkaya A, Bi C, Bradley AR, Christie CH, et al. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017;45:D271–81.
Jonić S. Cryo-electron microscopy analysis of structurally heterogeneous macromolecular complexes. Comput Struct Biotechnol J. 2016;14:385–90.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Advances in Neural Information Processing Systems 30. Long Beach, CA: USA; 2017. p 5998–6008.
Thomas N, Smidt T, Kearnes S, Yang L, Li L, Kohlhoff K, et al. Tensor field networks: rotation-and translation-equivariant neural networks for 3d point clouds. Computing Research Repository. 2018;abs/1802.08219:1–19.
Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Žídek A, et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–6.
Masrati G, Landau M, Ben-Tal N, Lupas A, Kosloff M, Kosinski J. Integrative structural biology in the era of accurate structure prediction. J Mol Biol. 2021;433:167127.
Cramer P. AlphaFold2 and the future of structural biology. Nat Struct Mol Biol. 2021;28:704–5.
Schiöth HB, Fredriksson R. The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gen Comp Endocrinol. 2005;142:94–101.
Isberg V, de Graaf C, Bortolato A, Cherezov V, Katritch V, Marshall FH, et al. Generic GPCR residue numbers—aligning topology maps while minding the gaps. Trends Pharmacol Sci. 2015;36:22–31.
Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–50.
Lu S, He X, Yang Z, Chai Z, Zhou S, Wang J, et al. Activation pathway of a G protein-coupled receptor uncovers conformational intermediates as targets for allosteric drug design. Nat Commun. 2021;12:4721.
Dror RO, Arlow DH, Maragakis P, Mildorf TJ, Pan AC, Xu H, et al. Activation mechanism of the β2-adrenergic receptor. Proc Natl Acad Sci USA. 2011;108:18684–9.
Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR dynamics: structures in motion. Chem Rev. 2016;117:139–55.
Ulloa-Aguirre A, Zariñán T, Jardón-Valadez E, Gutiérrez-Sagal R, Dias JA. Structure-function relationships of the follicle-stimulating hormone receptor. Front Endocrinol. 2018;29:707.
Wingler LM, Lefkowitz RJ. Conformational basis of G protein-coupled receptor signaling versatility. Trends Cell Biol. 2020;30:736–47.
Lu S, Zhang J. Small molecule allosteric modulators of G-protein-coupled receptors: drug–target Interactions. J Med Chem. 2019;62:24–45.
He X, Ni D, Lu S, Zhang J. Characteristics of allosteric proteins, sites, and modulators. In: Zhang J, Nussinov R, editors. Advances in experimental medicine and biology; v 1163. Protein Allostery in Drug Discovery. Singapore: Springer; 2019. p. 107–39.
Zhou Q, Yang D, Wu M, Guo Y, Guo W, Zhong L, et al. Common activation mechanism of class A GPCRs. Elife. 2019;8:e50279.
Mattedi G, Acosta-Gutiérrez S, Clark T, Gervasio FL. A combined activation mechanism for the glucagon receptor. Proc Natl Acad Sci USA. 2020;117:15414–22.
Fischer A, Smieško M, Sellner M, Lill MA. Decision making in structure-based drug discovery: visual inspection of docking results. J Med Chem. 2021;64:2489–500.
Jacobson KA. New paradigms in GPCR drug discovery. Biochem Pharmacol. 2015;98:541–55.
Roth BL, Irwin JJ, Shoichet BK. Discovery of new GPCR ligands to illuminate new biology. Nat Chem Biol. 2017;13:1143–51.
Liang YL, Belousoff MJ, Zhao P, Koole C, Fletcher MM, Truong TT, et al. Toward a structural understanding of class B GPCR peptide binding and activation. Mol Cell. 2020;77:656–68.
DeVree BT, Mahoney JP, Vélez-Ruiz GA, Rasmussen SGF, Kuszak AJ, Edwald E, et al. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature. 2016;535:182–6.
Weis WI, Kobilka BK. The molecular basis of G protein–coupled receptor activation. Annu Rev Biochem. 2018;87:897–919.
Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Madan BM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–94.
Josephs TM, Belousoff MJ, Liang YL, Piper SJ, Cao J, Garama DJ, et al. Structure and dynamics of the CGRP receptor in apo and peptide-bound forms. Science. 2021;372:eabf7258.
Tehan BG, Bortolato A, Blaney FE, Weir MP, Mason JS. Unifying family A GPCR theories of activation. Pharmacol Ther. 2014;143:51–60.
Munk C, Mutt E, Isberg V, Nikolajsen LF, Bibbe JM, Flock T, et al. An online resource for GPCR structure determination and analysis. Nat Methods. 2019;16:151–62.
Pándy-Szekeres G, Munk C, Tsonkov TM, Mordalski S, Harpsøe K, Hauser AS, et al. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 2018;46:D440–6.
Karageorgos V, Venihaki M, Sakellaris S, Pardalos M, Kontakis G, Matsoukas MT, et al. Current understanding of the structure and function of family B GPCRs to design novel drugs. Hormones. 2018;17:45–59.
Yang D, Zhou Q, Labroska V, Qin S, Darbalaei S, Wu Y, et al. G protein-coupled receptors: structure- and function-based drug discovery. Signal Transduct Target Ther. 2021;6:7.
Moore PB, Hendrickson WA, Henderson R, Brunger AT. The protein-folding problem: not yet solved. Science. 2022;375:507. https://doi.org/10.1126/science.abn9422.
Zhao S, Wu B, Stevens RC. Advancing chemokine GPCR structure based drug discovery. Structure. 2019;27:405–8.
Massink A, Amelia T, Karamychev A, IJzerman AP. Allosteric modulation of G protein-coupled receptors by amiloride and its derivatives. Perspectives for drug discovery? Med Res Rev. 2020;40:683–708.
Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, Katritch V, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–76.
Zhang X, He C, Wang M, Zhou Q, Yang D, Zhu Y, et al. Structures of the human cholecystokinin receptors bound to agonists and antagonists. Nat Chem Biol. 2021;17:1230–7.
McCorvy JD, Wacker D, Wang S, Agegnehu B, Liu J, Lansu K, et al. Structural determinants of 5-HT2B receptor activation and biased agonism. Nat Struct Mol Biol. 2018;25:787–96.
Kim K, Che T, Panova O, DiBerto JF, Lyu J, Krumm BE, et al. Structure of a hallucinogen-activated Gq-coupled 5-HT2A serotonin receptor. Cell. 2020;182:1574–88.
Rossmann MG. Molecular replacement-historical background. Acta Crystallogr D Biol Crystallogr. 2001;57:1360–6.
Acknowledgements
This work was partially supported by Lingang Laboratory grant (LG202102-01-01 to XC); Ministry of Science and Technology (China) grants (2018YFA0507002 to HEX); Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to HEX); Shanghai Municipal Science and Technology Major Project (HEX); CAS Strategic Priority Research Program (XDB37030103 to HEX); the National Natural Science Foundation of China (32130022 to HEX, 32171187 to YJ, 82121005 to HEX and YJ); and Shanghai Municipal Science and Technology Major Project.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
He, Xh., You, Cz., Jiang, Hl. et al. AlphaFold2 versus experimental structures: evaluation on G protein-coupled receptors. Acta Pharmacol Sin 44, 1–7 (2023). https://doi.org/10.1038/s41401-022-00938-y
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41401-022-00938-y
Keywords
This article is cited by
-
Barlow Twins deep neural network for advanced 1D drug–target interaction prediction
Journal of Cheminformatics (2025)
-
Research on the comprehensive dessert evaluation method in shale oil reservoirs based on fractal characteristics of conventional logging curves
Scientific Reports (2025)
-
Exploring the distinct activation mechanisms of neuromedin B receptor through multiple replica molecular dynamics simulations and Markov state modeling
Acta Pharmacologica Sinica (2025)
-
Exploring the constitutive activation mechanism of the class A orphan GPR20
Acta Pharmacologica Sinica (2025)
-
An update for AlphaFold3 versus experimental structures: assessing the precision of small molecule binding in GPCRs
Acta Pharmacologica Sinica (2025)