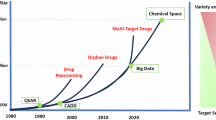
Abstract
The study of traditional medicine has garnered significant interest, resulting in various research areas including chemical composition analysis, pharmacological research, clinical application, and quality control. The abundance of available data has made databases increasingly essential for researchers to manage the vast amount of information and explore new drugs. In this article we provide a comprehensive overview and summary of 182 databases that are relevant to traditional medicine research, including 73 databases for chemical component analysis, 70 for pharmacology research, and 39 for clinical application and quality control from published literature (2000–2023). The review categorizes the databases by functionality, offering detailed information on websites and capacities to facilitate easier access. Moreover, this article outlines the primary function of each database, supplemented by case studies to aid in database selection. A practical test was conducted on 68 frequently used databases using keywords and functionalities, resulting in the identification of highlighted databases. This review serves as a reference for traditional medicine researchers to choose appropriate databases and also provides insights and considerations for the function and content design of future databases.

Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ikram RRR, Abd Ghani MK, Abdullah N. An analysis of application of health informatics in Traditional Medicine: A review of four Traditional Medicine Systems. Int J Med Inf. 2015;84:988–96.
Pan HQ, Yao CL, Yang WZ, Yao S, Huang Y, Zhang YB, et al. An enhanced strategy integrating offline two-dimensional separation and step-wise precursor ion list-based raster-mass defect filter: Characterization of indole alkaloids in five botanical origins of Uncariae Ramulus Cum Unicis as an exemplary application. J Chromatogr A. 2018;1563:124–34.
Chen XB, Yao CL, Hou JR, Nie M, Li Y, Wei WL, et al. Systematical characterization of gypenosides in Gynostemma pentaphyllum and the chemical composition variation of different origins. J Pharm Biomed Anal. 2023;232:115328.
Xu H, Li S, Liu J, Cheng J, Kang L, Li W, et al. Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19. Proc Natl Acad Sci USA. 2023;120:e2301775120.
Li XL, Zhang JQ, Li Y, Shen XJ, Yang HY, Yang L, et al. Medcheck: a novel software for automated de-formulation of traditional Chinese medicine (TCM) prescriptions by liquid chromatography-mass spectrometry. J Pharm Anal. 2024;14:100958.
Meng Q, Zhang J, Li X, Li Y, Shen X, Li Z, et al. ASAP-MS combined with mass spectrum similarity and binary code for rapid and intelligent authentication of 78 edible flowers. Food Chem. 2024;436:137776.
Wang C, Bi Q, Huang D, Wu S, Gao M, Li Y, et al. Identification of Pinelliae Rhizoma and its counterfeit species based on enzymatic signature peptides from toxic proteins. Phytomedicine. 2022;107:154451.
Li Z, Wu M, Wei W, An Y, Li Y, Wen Q, et al. Fingerprinting evaluation and Gut microbiota regulation of polysaccharides from Jujube (Ziziphus jujuba Mill.). Fruit Int J Mol Sci. 2023;24:7239.
Luo XX, Bi QR, Huang DD, Li Y, Yao CL, Zhang JQ, et al. Characterization of natural peptides in Pheretima by integrating proteogenomics and label-free peptidomics. J Pharm Anal. 2023;13:1070–9.
Hou JJ, Zhang ZJ, Zhang LL, Wu WY, Huang Y, Jia ZW, et al. Spatial lipidomics of eight edible nuts by desorption electrospray ionization with ion mobility mass spectrometry imaging. Food Chem. 2022;371:130893.
Shi X, Yang W, Qiu S, Hou J, Wu W, Guo D. Systematic profiling and comparison of the lipidomes from Panax ginseng, P. quinquefolius, and P. notoginseng by ultrahigh performance supercritical fluid chromatography/high-resolution mass spectrometry and ion mobility-derived collision cross section measurement. J Chromatogr A. 2018;1548:64–75.
Shen B. A new golden age of natural products drug discovery. Cell. 2015;163:1297–300.
Chi X, Wang S, Baloch Z, Zhang H, Li X, Zhang Z, et al. Research progress on classical traditional Chinese medicine formula Lily Bulb and Rehmannia Decoction in the treatment of depression. Biomed Pharmacother. 2019;112:108616.
Yu Y, Yao C, Guo DA. Insight into chemical basis of traditional Chinese medicine based on the state-of-the-art techniques of liquid chromatography−mass spectrometry. Acta Pharm Sin B. 2021;11:1469–92.
Liu X, Zhang JQ, Li Y, Yao CL, An YL, Wei WL, et al. In-depth profiling, nontargeted metabolomic and selective ion monitoring of eight chemical markers for simultaneous identification of different part of Eucommia ulmoides in 12 commercial products by UPLC/QDa. Food Chem. 2022;393:133346.
van Santen JA, Kautsar SA, Medema MH, Linington RG. Microbial natural product databases: moving forward in the multi-omics era. Nat Prod Rep. 2021;38:264–78.
Yu MX, Ma XQ, Song X, Huang YM, Jiang HT, Wang J, et al. Validation of the key active ingredients and anti-inflammatory and analgesic effects of Shenjin Huoxue mixture against Osteoarthritis by integrating network pharmacology approach and thin-layer Chromatography analysis. Drug Des Devel Ther. 2020;14:1145–56.
Huang W, Xie X, Huo L, Liang X, Wang X, Chen X. An integrative DNA barcoding framework of ladybird beetles (Coleoptera: Coccinellidae). Sci Rep. 2020;10:10063.
Sharma S, Joshi R, Kumar D. Metabolomics insights and bioprospection of Polygonatum verticillatum: An important dietary medicinal herb of alpine Himalaya. Food Res Int. 2021;148:110619.
Hou JJ, Zhang JQ, Yao CL, Bauer R, Khan IA, Wu WY, et al. Deeper chemical perceptions for better traditional Chinese medicine standards. Engineering. 2019;5:83–97.
Sorokina M, Steinbeck C. Review on natural products databases: where to find data in 2020. J Cheminform. 2020;12:20.
Medema MH. The year 2020 in natural product bioinformatics: an overview of the latest tools and databases11Electronic supplementary information (ESI) available. Nat Prod Rep. 2021;38:301–6.
Zhang R, Zhu X, Bai H, Ning K. Network pharmacology databases for traditional Chinese Medicine: Review and assessment. Front Pharmacol. 2019;10:123.
Zhao J, Yang J, Tian S, Zhang W. A survey of web resources and tools for the study of TCM network pharmacology. Quant Biol. 2019;7:17–29.
Blaženović I, Kind T, Ji J, Fiehn O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites. 2018;8:31.
Kim S, Chen J, Cheng TJ, Gindulyte A, He J, He SQ, et al. PubChem 2023 update. Nucleic Acids Res. 2023;51:D1373–80.
Pence HE, Williams A. An online chemical information resource. J Chem Educ. 2010;87:1123–4.
Mahmud S, Paul GK, Biswas S, Kazi T, Mahbub S, Mita MA, et al. Phytochemdb: a platform for virtual screening and computer-aided drug designing. Database (Oxf). 2022;2022:baac002.
Hastings J, Owen G, Dekker A, Ennis M, Kale N, Muthukrishnan V, et al. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44:D1214–9.
Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: A free tool to discover chemistry for biology. J Chem Inf Model. 2012;52:1757–68.
Chemical Structure Lookup Service (CSLS). Accessed 30 Dec. 2023. Available from: http://cactus.nci.nih.gov/lookup.
Yurekten O, Payne T, Tejera N, Amaladoss FX, Martin C, Williams M, et al. MetaboLights: open data repository for metabolomics. Nucleic Acids Res. 2023;52:D640–6.
Norsigian CJ, Pusarla N, McConn JL, Yurkovich JT, Dräger A, Palsson BO, et al. BiGG Models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res. 2020;48:D402–6.
Gu JY, Gui YS, Chen LR, Yuan G, Lu HZ, Xu XJ. Use of natural products as chemical library for drug discovery and network pharmacology. PLoS One. 2013;8:e62839.
Loub WD, Farnsworth NR, Soejarto DD, Quinn ML. Napralert-computer handing of natural product research data. J Chem Inf Comput Sci. 1985;25:99–103.
Miettinen K, Iñigo S, Kreft L, Pollier J, De Bo C, Botzki A, et al. The TriForC database: a comprehensive up-to-date resource of plant triterpene biosynthesis. Nucleic Acids Res. 2018;46:D586–94.
Chen CYC. TCM Database@Taiwan: The world’s largest traditional Chinese medicine database for drug screening in silico. PLoS One. 2011;6:e15939.
Novick PA, Ortiz OF, Poelman J, Abdulhay AY, Pande VS. SWEETLEAD: an in silico database of approved drugs, regulated chemicals, and herbal isolates for computer-aided drug discovery. PLoS One. 2013;8:e79568.
Rajoka MI, Idrees S, Khalid S, Ehsan B. Medherb: An interactive bioinformatics database and analysis resource for medicinally important herbs. Curr Bioinf. 2014;9:23–7.
Al Sharie AH, El-Elimat T, Al Zu’bi YO, Aleshawi AJ, Medina-Franco JL. Chemical space and diversity of seaweed metabolite database (SWMD): A cheminformatics study. J Mol Graph Model. 2020;100:107702.
Yue Y, Chu GX, Liu XS, Tang X, Wang W, Liu GJ, et al. TMDB: A literature-curated database for small molecular compounds found from tea. BMC Plant Biol. 2014;14:243.
Pathania S, Ramakrishnan SM, Bagler G. Phytochemica: a platform to explore phytochemicals of medicinal plants. Database (Oxf). 2015;2015:bav075.
Pathania S, Ramakrishnan SM, Randhawa V, Bagler G. SerpentinaDB: a database of plant-derived molecules of Rauvolfia serpentina. BMC Complement Alter Med. 2015;15:262.
Boonen J, Bronselaer A, Nielandt J, Veryser L, De Tré G, De Spiegeleer B. Alkamid database: Chemistry, occurrence and functionality of plant N-alkylamides. J Ethnopharmacol. 2012;142:563–90.
Yabuzaki J. Carotenoids Database: structures, chemical fingerprints and distribution among organisms. Database. 2017;2017:bax004.
Kim SK, Nam S, Jang H, Kim A, Lee JJ. TM-MC: a database of medicinal materials and chemical compounds in Northeast Asian traditional medicine. BMC Complement Alter Med. 2015;15:218.
Ntie-Kang F, Telukunta KK, Döring K, Sirnoben CV, Moumbock AFA, Malange YI, et al. NANPDB: A resource for natural products from Northern African sources. J Nat Prod. 2017;80:2067–76.
Simoben CV, Qaseem A, Moumbock AFA, Telukunta KK, Gunther S, Sippl W, et al. Pharmacoinformatic Investigation of medicinal plants from East Africa. Mol Inf. 2020;39:e2000163.
Ntie-Kang F, Onguéné PA, Fotso GW, Andrae-Marobela K, Bezabih M, Ndom JC, et al. Virtualizing the p-ANAPL library: A step towards drug discovery from African medicinal plants. PLoS One. 2014;9:e90655.
Ntie-Kang F, Zofou D, Babiaka SB, Meudom R, Scharfe M, Lifongo LL, et al. AfroDb: A select highly potent and diverse natural product library from African Medicinal Plants. PLoS One. 2013;8:e78085.
Ntie-Kang F, Mbah JA, Mbaze LM, Lifongo LL, Scharfe M, Hanna JN, et al. CamMedNP: Building the Cameroonian 3D structural natural products database for virtual screening. BMC Complement Alter Med. 2013;13:88.
Ntie-Kang F, Onguéné PA, Scharfe M, Owono LCO, Megnassan E, Mbaze LM, et al. ConMedNP: a natural product library from Central African medicinal plants for drug discovery. RSC Adv. 2014;4:409–19.
Diallo BN, Glenister M, Musyoka TM, Lobb K, Bishop Ö. SANCDB: an update on South African natural compounds and their readily available analogs. J Cheminform. 2021;13:37.
Meetei PA, Singh P, Nongdam P, Prabhu NP, Rathore R, Vindal V. NeMedPlant: a database of therapeutic applications and chemical constituents of medicinal plants from north-east region of India. Bioinformation. 2012;8:209–11.
Mirza SB, Bokhari H, Fatmi MQ. Exploring natural products from the biodiversity of pakistan for computational drug discovery studies: collection, optimization, design and development of a chemical database (ChemDP). Curr Comput-Aided Drug Des. 2015;11:102–9.
Pilón-Jiménez BA, Saldivar-Gonzalez FI, Diaz-Eufracio BI, Medina-Franco JL. BIOFACQUIM: A Mexican compound database of natural products. Biomolecules. 2019;9:31.
Nguyen-Vo TH, Le T, Pham D, Nguyen T, Le P, Nguyen A, et al. VIETHERB: A database for Vietnamese herbal species. J Chem Inf Model. 2019;59:1–9.
Mihaleva VV, te Beek TAH, van Zimmeren F, Moco S, Laatikainen R, Niemitz M, et al. MetIDB: A publicly accessible database of predicted and experimental 1H NMR Spectra of Flavonoids. Anal Chem. 2013;85:8700–7.
Fujita A, Aoki NP, Shinmachi D, Matsubara M, Tsuchiya S, Shiota M, et al. The international glycan repository GlyTouCan version 3.0. Nucleic Acids Res. 2021;49:D1529–33.
Yamada I, Hiraki AT, Miura N, Aoki-Kinoshita KF. GlycoNAVI: - GlycoAbun - ABUNDANCE RATIo of Glycans. Glycobiology. 2018;28:1067.
Frank M, Lütteke T, von der Lieth CW. GlycoMapsDB: a database of the accessible conformational space of glycosidic linkages. Nucleic Acids Res. 2007;35:287–90.
Kang X, Zhao WC, Widanage MDC, Kirui A, Ozdenvar U, Wang T. CCMRD: a solid-state NMR database for complex carbohydrates. J Biomol NMR. 2020;74:239–45.
Rojas-Macias MA, Mariethoz J, Andersson P, Jin CS, Venkatakrishnan V, Aoki NP, et al. Towards a standardized bioinformatics infrastructure for N- and O-glycomics. Nat Commun. 2019;10:3275.
Conroy MJ, Andrews RM, Andrews S, Cockayne L, Dennis EA, Fahy E, et al. LIPID MAPS: update to databases and tools for the lipidomics community. Nucleic Acids Res. 2023;52:D1677–82.
Foster JM, Moreno P, Fabregat A, Hermjakob H, Steinbeck C, Apweiler R, et al. LipidHome: A database of theoretical lipids optimized for high throughput mass spectrometry lipidomics. PLoS One. 2013;8:e61951.
Zhou ZW, Shen XT, Chen X, Tu J, Xiong X, Zhu ZJ. LipidIMMS Analyzer: integrating multi-dimensional information to support lipid identification in ion mobility-mass spectrometry based lipidomics. Bioinformatics. 2019;35:698–700.
Watanabe K, Yasugi E, Oshima M. How to search the glycolipid data in “LIPIDBANK for Web”, the newly developed lipid database in Japan. Trends Glycosci Glyc. 2000;12:175–84.
Aimo L, Liechti R, Hyka-Nouspikel N, Niknejad A, Gleizes A, Götz L, et al. The SwissLipids knowledgebase for lipid biology. Bioinformatics. 2015;31:2860–6.
Kuo TC, Tseng YJ. LipidPedia: a comprehensive lipid knowledgebase. Bioinformatics. 2018;34:2982–7.
Bateman A, Martin MJ, Orchard S, Magrane M, Ahmad S, Alpi E, et al. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023;51:D523–D31.
Liu ZD, Li L, Xue B, Zhao DD, Zhang YL, Yan XF. A new lectin from Auricularia auricula inhibited the proliferation of lung cancer cells and improved pulmonary flora. BioMed Res Int. 2021;2021:5597135.
Burley SK, Bhikadiya C, Bi CX, Bittrich S, Chao HY, Chen L, et al. RCSB Protein Data Bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023;51:D488–508.
López-Pérez JL, Therón R, del Olmo E, Díaz D. NAPROC-13: a database for the dereplication of natural product mixtures in bioassay-guided protocols. Bioinformatics. 2007;23:3256–7.
Glisic S, Sencanski M, Perovic V, Stevanovic S, García-Sosa AT. Arginase Flavonoid Anti-Leishmanial in silico inhibitors flagged against anti-targets. Molecules. 2016;21:589.
Tulpan D, Léger S, Belliveau L, Culf A, Cuperlovic-Culf M. MetaboHunter: an automatic approach for identification of metabolites from 1H-NMR spectra of complex mixtures. BMC Bioinform. 2011;12:400.
Wishart DS, Sayeeda Z, Budinski Z, Guo AC, Lee BL, Berjanskii M, et al. NP-MRD: the natural products magnetic resonance database. Nucleic Acids Res. 2022;50:D665–77.
Steinbeck C, Kuhn S. NMRShiftDB - compound identification and structure elucidation support through a free community-built web database. Phytochemistry. 2004;65:2711–7.
Pilon AC, Valli M, Dametto AC, Pinto MEF, Freire RT, Castro-Gamboa I, et al. NuBBEDB: an updated database to uncover chemical and biological information from Brazilian biodiversity. Sci Rep. 2017;7:7215.
Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–8.
Clendinen CS, Pasquel C, Ajredini R, Edison AS. 13C NMR metabolomics: inadequate network analysis. Anal Chem. 2015;87:5698–706.
Fischedick JT, Johnson SR, Ketchum REB, Croteau RB, Lange BM. NMR spectroscopic search module for Spektraris, an online resource for plant natural product identification - Taxane diterpenoids from Taxus x media cell suspension cultures as a case study. Phytochemistry. 2015;113:87–95.
Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45:703–14.
Mistrik R. mzCLOUD: A spectral tree library for the Identification of “unknown unknowns”. Abstr Pap Am Chem Soc. 2018;255:31.
Xue JC, Guijas C, Benton HP, Warth B, Siuzdak G. METLIN MS2 molecular standards database: a broad chemical and biological resource. Nat Meth. 2020;17:953–4.
Song C, Zhang Y, Manzoor MA, Li G. Identification of alkaloids and related intermediates of Dendrobium officinale by solid-phase extraction coupled with high-performance liquid chromatography tandem mass spectrometry. Front Plant Sci. 2022;13:952051.
Nothias LF, Petras D, Schmid R, Dührkop K, Rainer J, Sarvepalli A, et al. Feature-based molecular networking in the GNPS analysis environment. Nat Meth. 2020;17:905–8.
Xie HF, Kong YS, Li RZ, Nothias LF, Melnik AV, Zhang H, et al. Feature-based molecular networking analysis of the metabolites produced by in vitro solid-state fermentation reveals pathways for the bioconversion of epigallocatechin gallate. J Agric Food Chem. 2020;68:7995–8007.
Nakamura Y, Afendi FM, Parvin AK, Ono N, Tanaka K, Morita AH, et al. KNApSAcK metabolite activity database for retrieving the relationships between metabolites and biological activities. Plant Cell Physiol. 2014;55:e7.
Sawada Y, Nakabayashi R, Yamada Y, Suzuki M, Sato M, Sakata A, et al. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: A plant-specific MS/MS-based data resource and database. Phytochemistry. 2012;82:38–45.
Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, et al. GMD@CSB.DB:: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–8.
Hayes CA, Karlsson NG, Struwe WB, Lisacek F, Rudd PM, Packer NH, et al. UniCarb-DB: a database resource for glycomic discovery. Bioinformatics. 2011;27:1343–4.
Wishart DS, Guo AC, Oler E, Wang F, Anjum A, Peters H, et al. HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Res. 2022;50:D622–31.
Han YQ, Guo WJ, Li XX, Xu XH, Yang JX, Xie SX, et al. LC-MS-based metabolomics reveals the in vivo effect of Shegan Mahuang Decoction in an OVA-induced rat model of airway hyperresponsiveness. Mol Omics. 2022;18:957–66.
Dabb S, Blunt J, Munro M. MarinLit: Database and essential tools for the marine natural products community. Abstr Pap Am Chem Soc. 2014;248:25.
Brkljaca R, Göker ES, Urban S. Dereplication and chemotaxonomical studies of marine algae of the Ochrophyta and Rhodophyta phyla. Mar Drugs. 2015;13:2714–31.
Tanabe K, Hayamizu K, Ono S, Wasada N, Someno K, Nonaka S, et al. Spectral database system SDBS On PC with CD-ROM. Anal Sci. 1991;7:711–2.
Seiler KP, George GA, Happ MP, Bodycombe NE, Carrinski HA, Norton S, et al. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36:D351–9.
Maeda MH, Kondo K. Three-Dimensional Structure Database of Natural Metabolites (3DMET): a novel database of curated 3D structures. J Chem Inf Model. 2013;53:527–33.
Zhao H, Yang Y, Wang SQ, Yang X, Zhou KC, Xu CL, et al. NPASS database update 2023: quantitative natural product activity and species source database for biomedical research. Nucleic Acids Res. 2023;51:D621–8.
Ibrahim MAA, Abdeljawaad KAA, Abdelrahman AHM, Jaragh-Alhadad LA, Oraby HF, Elkaeed EB, et al. Exploring natural product activity and species source candidates for hunting ABCB1 transporter inhibitors: an in silico drug discovery study. Molecules. 2022;27:3104.
Gradinaru TC, Petran M, Dragos D, Gilca M. PlantMolecularTasteDB: a database of taste active phytochemicals. Front Pharmacol. 2022;12:751712.
Sharma A, Dutta P, Sharma M, Rajput NK, Dodiya B, Georrge JJ, et al. BioPhytMol: a drug discovery community resource on anti-mycobacterial phytomolecules and plant extracts. J Cheminform. 2014;6:46.
Choi H, Cho SY, Pak HJ, Kim Y, Choi JY, Lee YJ, et al. NPCARE: database of natural products and fractional extracts for cancer regulation. J Cheminform. 2017;9:2.
Tan XJ, Fu JH, Yuan ZX, Zhu LJ, Fu LL. ACNPD: The database for elucidating the relationships between natural products, compounds, molecular mechanisms, and cancer types. Front Pharmacol. 2021;12:746067.
Ibezim A, Debnath B, Ntie-Kang F, Mbah CJ, Nwodo NJ. Binding of anti-Trypanosoma natural products from African flora against selected drug targets: a docking study. Med Chem Res. 2017;26:562–79.
Lin YC, Wang CC, Chen IS, Jheng JL, Li JH, Tung CW. TIPdb: A database of anticancer, antiplatelet, and antituberculosis phytochemicals from indigenous plants in Taiwan. Sci World J. 2013;2013:736386.
Ntie-Kang F, Nwodo JN, Ibezim A, Simoben CV, Karaman B, Ngwa VF, et al. Molecular modeling of potential anticancer agents from African medicinal plants. J Chem Inf Model. 2014;54:2433–50.
Vetrivel U, Subramanian N, Pilla K. InPACdb–Indian plant anticancer compounds database. Bioinformation. 2009;4:71–4.
Kang H, Tang KL, Liu Q, Sun Y, Huang Q, Zhu RX, et al. HIM-herbal ingredients in-vivo metabolism database. J Cheminform. 2013;5:28.
Wang GS, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:D1087–93.
Gogoladze G, Grigolava M, Vishnepolsky B, Chubinidze M, Duroux P, Lefranc MP, et al. DBAASP: database of antimicrobial activity and structure of peptides. FEMS Microbiol Lett. 2014;357:63–8.
Shi GB, Kang XY, Dong FY, Liu YC, Zhu N, Hu YX, et al. DRAMP 3.0: an enhanced comprehensive data repository of antimicrobial peptides. Nucleic Acids Res. 2022;50:D488–96.
Waghu FH, Gopi L, Barai RS, Ramteke P, Nizami B, Idicula-Thomas S. CAMP: Collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res. 2014;42:D1154–8.
Jhong JH, Chi YH, Li WC, Lin TH, Huang KY, Lee TY. dbAMP: an integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. 2019;47:D285–97.
Feurstein C, Meyer V, Jung S. Structure–activity predictions from computational mining of protein databases to assist modular design of antimicrobial peptides. Front Microbiol. 2022;13:812903.
Wang J, Yin TL, Xiao XW, He D, Xue ZD, Jiang XN, et al. StraPep: a structure database of bioactive peptides. Database (Oxf). 2018;2018:bay038.
Shtatland T, Guettler D, Kossodo M, Pivovarov M, Weissleder R. PepBank - a database of peptides based on sequence text mining and public peptide data sources. BMC Bioinform. 2007;8:280.
Agrawal P, Bhalla S, Usmani SS, Singh S, Chaudhary K, Raghava GPS, et al. CPPsite 2.0: a repository of experimentally validated cell-penetrating peptides. Nucleic Acids Res. 2016;44:D1098–103.
Jaiswal M, Singh A, Kumar S. PTPAMP: prediction tool for plant-derived antimicrobial peptides. Amino Acids. 2023;55:1–17.
Lin J, Wen L, Zhou Y, Wang S, Ye H, Su J, et al. PepQSAR: a comprehensive data source and information platform for peptide quantitative structure–activity relationships. Amino Acids. 2023;55:235–42.
Das D, Jaiswal M, Khan FN, Ahamad S, Kumar S. PlantPepDB: A manually curated plant peptide database. Sci Rep. 2020;10:2194.
Miller MA, Hazard GF, Hudson VW, Hilt C, Fang J, Mayer D, et al. ChemIDplus: A free, web-based portal to a variety of compound-based information. Abstr Pap Am Chem Soc. 2003;226:U305.
Schmidt U, Struck S, Gruening B, Hossbach J, Jaeger IS, Parol R, et al. SuperToxic: a comprehensive database of toxic compounds. Nucleic Acids Res. 2009;37:D295–9.
Günthardt BF, Hollender J, Hungerbühler K, Scheringer M, Bucheli TD. Comprehensive toxic plants-phytotoxins database and its application in assessing aquatic micropollution potential. J Agric Food Chem. 2018;66:7577–88.
He QY, He QZ, Deng XC, Yao L, Meng E, Liu ZH, et al. ATDB: a uni-database platform for animal toxins. Nucleic Acids Res. 2008;36:D293–7.
Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu XD, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell. 2017;171:1437–52.
Gu Y, Huang P, Cheng T, Yang J, Wu G, Sun Y, et al. A multiomics and network pharmacological study reveals the neuroprotective efficacy of Fu-Fang-Dan-Zhi tablets against glutamate-induced oxidative cell death. Comput Biol Med. 2022;148:105873.
Jiang HM, Hu C, Chen MJ. The advantages of connectivity map applied in Traditional Chinese Medicine. Front Pharmacol. 2021;12:474267.
Tian S, Zhang JB, Yuan SL, Wang Q, Lv C, Wang JX, et al. Exploring pharmacological active ingredients of traditional Chinese medicine by pharmacotranscriptomic map in ITCM. Brief Bioinform. 2023;24:bbad027.
Safran M, Dalah I, Alexander J, Rosen N, Stein TI, Shmoish M, et al. GeneCards Version 3: the human gene integrator. Database (Oxf). 2010;2010:baq020.
Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–55.
Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online mendelian inheritance in man (OMIM). Hum Mutat. 2000;15:57–61.
Mu BX, Li YX, Ye NY, Liu SL, Zou X, Qian J, et al. Understanding apoptotic induction by Sargentodoxa cuneata-Patrinia villosa herb pair via PI3K/AKT/mTOR signalling in colorectal cancer cells using network pharmacology and cellular studies. J Ethnopharmacol. 2024;319:117342.
Gilson MK, Liu TQ, Baitaluk M, Nicola G, Hwang L, Chong J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–53.
Wang RX, Fang XL, Lu YP, Yang CY, Wang SM. The PDBbind database: Methodologies and updates. J Med Chem. 2005;48:4111–9.
Placzek S, Schomburg I, Chang A, Jeske L, Ulbrich M, Tillack J, et al. BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res. 2017;45:D380–8.
Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, et al. A draft map of the human proteome. Nature. 2014;509:575–81.
Harini K, Srivastava A, Kulandaisamy A, Gromiha MM. ProNAB: database for binding affinities of protein-nucleic acid complexes and their mutants. Nucleic Acids Res. 2022;50:D1528–34.
Zhou Y, Zhang YT, Zhao DH, Yu XY, Shen XY, Zhou Y, et al. TTD: Therapeutic Target Database describing target druggability information. Nucleic Acids Res. 2023;52:D1465–77.
Mendez D, Gaulton A, Bento AP, Chambers J, De Veij M, Félix E, et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019;47:D930–40.
Knox C, Wilson M, Klinger CM, Franklin M, Oler E, Wilson A, et al. DrugBank 6.0: the DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2023;52:D1265–D75.
Gao ZT, Li HL, Zhang HL, Liu XF, Kang L, Luo XM, et al. PDTD: a web-accessible protein database for drug target identification. BMC Bioinform. 2008;9:104.
Yan DY, Zheng GH, Wang CC, Chen ZK, Mao TT, Gao J, et al. HIT 2.0: an enhanced platform for Herbal Ingredients’ Targets. Nucleic Acids Res. 2022;50:D1238–D43.
Mangal M, Sagar P, Singh H, Raghava GPS, Agarwal SM. NPACT: naturally occurring plant-based anti-cancer compound-activity-target database. Nucleic Acids Res. 2013;41:D1124–9.
Huang L, Xie DL, Yu YR, Liu HL, Shi Y, Shi TL, et al. TCMID 2.0: a comprehensive resource for TCM. Nucleic Acids Res. 2018;46:D1117–20.
Bultum LE, Woyessa AM, Lee D. ETM-DB: integrated Ethiopian traditional herbal medicine and phytochemicals database. BMC Complement Alter Med 2019;19:212.
Tao WY, Li BH, Gao S, Bai YF, Shar PA, Zhang WJ, et al. CancerHSP: anticancer herbs database of systems pharmacology. Sci Rep. 2015;5:11481.
Gu JY, Gui YS, Chen LR, Yuan G, Xu XJ. CVDHD: a cardiovascular disease herbal database for drug discovery and network pharmacology. J Cheminform. 2013;5:51.
Choi W, Choi CH, Kim YR, Kim SJ, Na CS, Lee H. HerDing: herb recommendation system to treat diseases using genes and chemicals. Database. 2016;2016:baw011.
Li X, Ren J, Zhang W, Zhang ZM, Yu JC, Wu JW, et al. LTM-TCM: A comprehensive database for the linking of Traditional Chinese Medicine with modern medicine at molecular and phenotypic levels. Pharm Res. 2022;178:106185.
Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44:D380–4.
Kong XR, Liu C, Zhang ZZ, Cheng MQ, Mei ZJ, Li XD, et al. BATMAN-TCM 2.0: an enhanced integrative database for known and predicted interactions between traditional Chinese medicine ingredients and target proteins. Nucleic Acids Res. 2023;52:D1110–20.
Zhang YQ, Li X, Shi YL, Chen T, Xu ZJ, Wang P, et al. ETCM v2.0: An update with comprehensive resource and rich annotations for traditional Chinese medicine. Acta Pharma Sin B. 2023;13:2559–71.
Lee AY, Park W, Kang TW, Cha MH, Chun JM. Network pharmacology-based prediction of active compounds and molecular targets in Yijin-Tang acting on hyperlipidaemia and atherosclerosis. J Ethnopharmacol. 2018;221:151–9.
Huang YT, Zhang K, Wang X, Guo KM, Li XQ, Chen F, et al. Multi-omics approach for identification of molecular alterations of QiShenYiQi dripping pills in heart failure with preserved ejection fraction. J Ethnopharmacol. 2023;315:116673.
Milacic M, Beavers D, Conley P, Gong CQ, Gillespie M, Griss J, et al. The reactome pathway knowledgebase 2024. Nucleic Acids Res. 2023;52:D672–8.
Varusai TM, Jupe S, Sevilla C, Matthews L, Gillespie M, Stein L, et al. Using reactome to build an autophagy mechanism knowledgebase. Autophagy. 2021;17:1543–54.
Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–46.
Blake JA, Christie KR, Dolan ME, Drabkin HJ, Hill DP, Ni L, et al. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–56.
Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30.
Zhuang JM, Mo JH, Huang ZN, Yan YL, Wang ZY, Cao XQ, et al. Mechanisms of Xiaozheng decoction for anti-bladder cancer effects via affecting the GSK3β/β-catenin signaling pathways: a network pharmacology-directed experimental investigation. Chin Med. 2023;18:104.
Jewison T, Su YL, Disfany FM, Liang YJ, Knox C, Maciejewski A, et al. SMPDB 2.0: big improvements to the small molecule pathway database. Nucleic Acids Res. 2014;42:D478–84.
Caspi R, Billington R, Keseler IM, Kothari A, Krummenacker M, Midford PE, et al. The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 2020;48:D445–53.
Davis AP, Wiegers TC, Johnson RJ, Sciaky D, Wiegers J, Mattingly CJ. Comparative Toxicogenomics Database (CTD): update 2023. Nucleic Acids Res. 2023;51:D1257–62.
Banerjee P, Erehman J, Gohlke BO, Wilhelm T, Preissner R, Dunkel M. Super Natural II-a database of natural products. Nucleic Acids Res. 2015;43:D935–D9.
Fang YC, Huang HC, Chen HH, Juan HF. TCMGeneDIT: a database for associated traditional Chinese medicine, gene and disease information using text mining. BMC Complement Alter Med. 2008;8:58.
Hou DY, Lin HB, Feng YH, Zhou KC, Li XX, Yang Y, et al. CMAUP database update 2024: extended functional and association information of useful plants for biomedical research. Nucleic Acids Res. 2023;52:D1508–18.
Fang SS, Dong L, Liu L, Guo JC, Zhao LH, Zhang JY, et al. HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 2021;49:D1197–206.
Ru JL, Li P, Wang JN, Zhou W, Li BH, Huang C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13.
Lv QJ, Chen GX, He HH, Yang ZD, Zhao L, Chen HY, et al. TCMBank: bridges between the largest herbal medicines, chemical ingredients, target proteins, and associated diseases with intelligence text mining. Chem Sci. 2023;14:10684–701.
Wu Y, Zhang FL, Yang K, Fang SS, Bu DC, Li H, et al. SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 2019;47:D1110–7.
Liu ZH, Cai CP, Du JW, Liu BD, Cui L, Fan XD, et al. TCMIO: A comprehensive database of traditional chinese medicine on immuno-oncology. Front Pharmacol. 2020;11:439.
Li BQ, Ma CF, Zhao XY, Hu ZG, Du TF, Xu XM, et al. YaTCM: Yet another traditional Chinese medicine database for drug discovery. Comput Struct Biotechnol J. 2018;16:600–10.
Zhang RZ, Yu SJ, Bai H, Ning K. TCM-Mesh: The database and analytical system for network pharmacology analysis for TCM preparations. Sci Rep. 2017;7:2821.
Chen X, Zhou H, Liu YB, Wang JF, Li H, Ung CY, et al. Database of traditional Chinese medicine and its application to studies of mechanism and to prescription validation. Br J Pharmacol. 2006;149:1092–103.
Huang J, Zheng YX, Wu WX, Xie T, Yao H, Pang XB, et al. CEMTDD: The database for elucidating the relationships among herbs, compounds, targets and related diseases for Chinese ethnic minority traditional drugs. Oncotarget. 2015;6:17675–84.
Chowdhury S, Sarkar RR. Comparison of human cell signaling pathway databases—evolution, drawbacks and challenges. Database. 2015;2015:bau126.
Barbarino JM, Whirl-Carrillo M, Altman RB, Klein TE. PharmGKB: A worldwide resource for pharmacogenomic information. Wiley Interdiscip Rev Syst Biol Med. 2018;10:e1417.
Zhang Y, Man Ip C, Lai YS, Zuo Z. Overview of current herb-drug interaction databases. Drug Metab Dispos. 2022;50:86–94.
Wang LL, Tafjord O, Cohan A, Jain S, Skjonsberg S, Schoenick C, et al. SUPP.AI: finding evidence for supplement-drug interactions. In: Association for Computational Linguistics. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics: System Demonstrations; Online; 2020. p 362–71.
Birer-Williams C, Gufford BT, Chou ER, Alilio M, VanAlstine S, Morley RE, et al. A new data repository for pharmacokinetic natural product-drug interactions: from chemical characterization to clinical studies. Drug Metab Dispos. 2020;48:1104–12.
Hachad H, Ragueneau-Majlessi I, Levy RH. Management of drug interactions of new drugs in multicenter trials using the metabolism and transport drug interaction database. Annual Meeting of the American-Association-of-Pharmaceutical-Scientists; 2007 Nov; San Diego, USA. Switzerland: Springer; 2010.
Zhang Y, Wang N, Du X, Chen T, Yu Z, Qin Y, et al. SoFDA: an integrated web platform from syndrome ontology to network-based evaluation of disease–syndrome–formula associations for precision medicine. Sci Bull. 2022;67:1097–101.
Fang XL, Shao L, Zhang H, Wang SM. CHMIS-C: A comprehensive herbal medicine information system for cancer. J Med Chem. 2005;48:1481–8.
Vivek-Ananth RP, Mohanraj K, Sahoo AK, Samal A. IMPPAT 2.0: an enhanced and expanded phytochemical atlas of Indian medicinal plants. Acs Omega. 2023;8:8827–45.
Ratnasingham S, Hebert PDN. Bold: The Barcode of life data system. Mol Ecol Notes. 2007;7:355–64.
Liao BS, Hu HY, Xiao SM, Zhou GR, Sun W, Chu Y, et al. Global Pharmacopoeia Genome Database is an integrated and mineable genomic database for traditional medicines derived from eight international pharmacopoeias. Sci China Life Sci. 2022;65:809–17.
Wong TH, But GWC, Wu HY, Tsang SSK, Lau DTW, Shaw PC. Medicinal Materials DNA Barcode Database (MMDBD) version 1.5-one-stop solution for storage, BLAST, alignment and primer design. Database (Oxf). 2018;2018:bay112.
Chen SL, Pang XH, Song JY, Shi LC, Yao H, Han JP, et al. A renaissance in herbal medicine identification: From morphology to DNA. Biotechnol Adv. 2014;32:1237–44.
Kim S, Kim CB, Min GS, Suh Y, Bhak J, Woo T, et al. Korea Barcode of Life Database System (KBOL). Anim Cells Syst. 2012;16:11–9.
Lim J, Kim SY, Kim S, Eo HS, Kim CB, Paek WK, et al. BioBarcode: a general DNA barcoding database and server platform for Asian biodiversity resources. BMC Genomics. 2009;10:S8.
Meng F, Tang Q, Chu T, Li X, Lin Y, Song X, et al. TCMPG: an integrative database for traditional Chinese medicine plant genomes. Hortic Res. 2022;9:uhac060.
Sayers EW, Cavanaugh M, Clark K, Pruitt KD, Sherry ST, Yankie L, et al. GenBank 2023 update. Nucleic Acids Res. 2023;51:D141–4.
Lv YN, Yang CY, She LC, Zhang ZL, Xu AS, Zhang LX, et al. Identification of medicinal plants within the Apocynaceae family using ITS2 and psbA-trnH barcodes. Chin J Nat Med. 2020;18:594–605.
Robbertse B, Strope PK, Chaverri P, Gazis R, Ciufo S, Domrachev M, et al. Improving taxonomic accuracy for fungi in public sequence databases: applying ‘one name one species’ in well-defined genera with Trichoderma/Hypocrea as a test case. Database. 2017;2017:bax072.
Li WJ, O’Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, et al. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021;49:D1020–8.
Harrison PW, Amode MR, Austine-Orimoloye O, Azov AG, Barba M, Barnes I, et al. Ensembl 2024. Nucleic Acids Res. 2023;52:D891–9.
Bolser DM, Kerhornou A, Walts B, Kersey P. Triticeae resources in Ensembl plants. Plant Cell Physiol. 2015;56:e3.
Xue YB, Bao YM, Zhang Z, Zhao WM, Xiao JF, He SM, et al. Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2023. Nucleic Acids Res. 2023;51:D18–28.
Carpenter EJ, Matasci N, Ayyampalayam S, Wu SX, Sun J, Yu J, et al. Access to RNA-sequencing data from 1173 plant species: The 1000 Plant transcriptomes initiative (1KP). GigaScience. 2019;8:giz126.
Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49:W29–35.
Terlouw BR, Blin K, Navarro-Muñoz JC, Avalon NE, Chevrette MG, Egbert S, et al. MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 2023;51:D603–10.
Ren L, Xu Y, Ning L, Pan X, Li Y, Zhao Q, et al. TCM2COVID: A resource of anti-COVID-19 traditional Chinese medicine with effects and mechanisms. Imeta. 2022;1:e42.
Tao QY, Du JX, Li XT, Zeng JY, Tan B, Xu JH, et al. Network pharmacology and molecular docking analysis on molecular targets and mechanisms of Huashi Baidu formula in the treatment of COVID-19. Drug Dev Ind Pharmacol. 2020;46:1345–53.
Zhu YW, Yan XF, Ye TJ, Hu J, Wang XL, Qiu FJ, et al. Analyzing the potential therapeutic mechanism of Huashi Baidu Decoction on severe COVID-19 through integrating network pharmacological methods. J Tradit Complement Med. 2021;11:180–7.
Nie JX, Qian WX, Wang MS, Wang JY, Zhang Y, Liu HM, et al. Research status and comparative study on toxicity related databases of traditional Chinese medicine. Zhong Cao Yao. 2023;54:7588–96.
Draper J, Enot DP, Parker D, Beckmann M, Snowdon S, Lin WC, et al. Metabolite signal identification in accurate mass metabolomics data with MZedDB, an interactive m/z annotation tool utilising predicted ionisation behaviour ‘rules’. BMC Bioinform. 2009;10:227.
Yang Y, Zhou D, Zhang X, Shi Y, Han J, Zhou L, et al. D3AI-CoV: a deep learning platform for predicting drug targets and for virtual screening against COVID-19. Brief Bioinform. 2022;23:bbac147.
Acknowledgements
The work was supported by the Qi-Huang Chief Scientist Project of the National Administration of Traditional Chinese Medicine (2020), the Sanming Project of Medicine in Shenzhen (SZZYSM202106004), Key Program of National Natural Science Foundation of China (82130111), National Natural Science Foundation of China (82003940), Key-Area Research and Development Program of Guangdong Province (2020B1111110007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Xl., Zhang, Jq., Shen, Xj. et al. Overview and limitations of database in global traditional medicines: A narrative review. Acta Pharmacol Sin 46, 235–263 (2025). https://doi.org/10.1038/s41401-024-01353-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41401-024-01353-1