Abstract
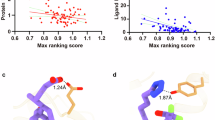
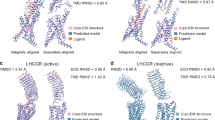
G protein-coupled receptors (GPCRs) are critical drug targets involved in numerous physiological processes, yet many of their structures remain unresolved due to inherent flexibility and diverse ligand interactions. This study systematically evaluates the accuracy of AlphaFold3-predicted GPCR structures compared to experimentally determined structures, with a primary focus on ligand-bound states. Our analysis reveals that while AlphaFold3 shows improved performance over AlphaFold2 in predicting overall GPCR backbone architecture, significant discrepancies persist in ligand-binding poses, particularly for ions, peptides, and proteins. Despite advancements, these limitations constrain the utility of AlphaFold3 models in functional studies and structure-based drug design, where high-resolution details of ligand interactions are crucial. We assess the accuracy of predicted structures across various ligand types, quantifying deviations in binding pocket geometries and ligand orientations. Our findings highlight specific challenges in the computational prediction of ligand-bound GPCR structures, emphasizing areas where further refinement is needed. This study provides valuable insights for researchers using AlphaFold3 in GPCR studies, underscores the ongoing necessity for experimental structure determination, and offers direction for improving protein–ligand interaction predictions in future computational models.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–d44.
Yang Z, Zeng X, Zhao Y, Chen R. AlphaFold2 and its applications in the fields of biology and medicine. Signal Transduct Target Ther 2023;8:115.
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500.
He XH, You CZ, Jiang HL, Jiang Y, Xu HE, Cheng X. AlphaFold2 versus experimental structures: evaluation on G protein-coupled receptors. Acta Pharmacol Sin. 2023;44:1–7.
Duan J, He XH, Li SJ, Xu HE. Cryo-electron microscopy for GPCR research and drug discovery in endocrinology and metabolism. Nat Rev Endocrinol. 2024;20:349–65.
Suno R. Exploring diverse signaling mechanisms of G protein-coupled receptors through structural biology. J Biochem. 2024;175:357–65.
Lu S, He X, Yang Z, Chai Z, Zhou S, Wang J, et al. Activation pathway of a G protein-coupled receptor uncovers conformational intermediates as targets for allosteric drug design. Nat Commun. 2021;12:4721.
Guo L, Zhang Y, Fang G, Tie L, Zhuang Y, Xue C, et al. Ligand recognition and G protein coupling of the human itch receptor MRGPRX1. Nat Commun. 2023;14:5004.
Betke KM, Wells CA, Hamm HE. GPCR mediated regulation of synaptic transmission. Prog Neurobiol. 2012;96:304–21.
He X, Duan J, Ji Y, Zhao L, Jiang H, Jiang Y, et al. Hinge region mediates signal transmission of luteinizing hormone and chorionic gonadotropin receptor. Comput Struct Biotechnol J. 2022;20:6503–11.
Duan J, Xu P, Luan X, Ji Y, He X, Song N, et al. Hormone- and antibody-mediated activation of the thyrotropin receptor. Nature. 2022;609:854–9.
Liu H, Zhang Q, He X, Jiang M, Wang S, Yan X, et al. Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84. Nat Commun. 2023;14:3271.
Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–42.
Zhang M, Chen T, Lu X, Lan X, Chen Z, Lu S. G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery. Signal Transduct Target Ther. 2024;9:88.
Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR dynamics: structures in motion. Chem Rev. 2017;117:139–55.
Liu H, Guo S, Dai A, Xu P, Li X, Huang S, et al. Structural and functional evidence that GPR30 is not a direct estrogen receptor. Cell Res. 2024;34:530–3.
Zhuang Y, Wang Y, He B, He X, Zhou XE, Guo S, et al. Molecular recognition of morphine and fentanyl by the human μ-opioid receptor. Cell. 2022;185:4361–75. e19.
He X, Huang N, Qiu Y, Zhang J, Liu Y, Yin XL, et al. Conformational selection mechanism provides structural insights into the optimization of APC-Asef inhibitors. Molecules. 2021;26:962.
Pándy-Szekeres G, Caroli J, Mamyrbekov A, Kermani AA, Keserű GM, Kooistra AJ, et al. GPCRdb in 2023: state-specific structure models using AlphaFold2 and new ligand resources. Nucleic Acids Res. 2023;51:D395–d402.
Shimada I, Ueda T, Kofuku Y, Eddy MT, Wüthrich K. GPCR drug discovery: integrating solution NMR data with crystal and cryo-EM structures. Nat Rev Drug Discov. 2019;18:59–82.
Fan W, Xu Y, He X, Luo P, Zhu J, Li J, et al. Molecular basis for the activation of PAF receptor by PAF. Cell Rep. 2024;43:114422.
Shen S, Wang D, Liu H, He X, Cao Y, Chen J, et al. Structural basis for hormone recognition and distinctive Gq protein coupling by the kisspeptin receptor. Cell Rep. 2024;43:114389.
He X, Ni D, Lu S, Zhang J. Characteristics of allosteric proteins, sites, and modulators. Adv Exp Med Biol. 2019;1163:107–39.
Ciancetta A, Jacobson KA. Breakthrough in GPCR crystallography and its impact on computer-aided drug design. Methods Mol Biol. 2018;1705:45–72.
Duan J, Liu H, Zhao F, Yuan Q, Ji Y, Cai X, et al. GPCR activation and GRK2 assembly by a biased intracellular agonist. Nature. 2023;620:676–81.
Mafi A, Kim SK, Goddard WA 3rd. The mechanism for ligand activation of the GPCR-G protein complex. Proc Natl Acad Sci USA. 2022;119:e2110085119.
Corso G, Stärk H, Jing B, Barzilay R, Jaakkola T. Diffdock: diffusion steps, twists, and turns for molecular docking. In: International Conference on Learning Representations. Kigali, Rwanda: Machine Learning for Sciences; 2023.
Su M, Yang Q, Du Y, Feng G, Liu Z, Li Y, et al. Comparative assessment of scoring functions: the CASF-2016 update. J Chem Inf Model. 2019;59:895–913.
Goetz L, Seedat N, Vandersluis R, van der Schaar M. Generalization—a key challenge for responsible AI in patient-facing clinical applications. npj Digit Med. 2024;7:126.
Huisman M, Hannink G. The AI generalization gap: one size does not fit all. Radiol Artif Intell. 2023;5:e230246.
Chakravarty D, Schafer JW, Chen EA, Thole JF, Ronish LA, Lee M, et al. AlphaFold predictions of fold-switched conformations are driven by structure memorization. Nat Commun. 2024;15:7296.
Jing B, Berger B, Jaakkola T. AlphaFold meets flow matching for generating protein ensembles. In: International Conference on Machine Learning. Vienna, Austria; 2024.
Li S, Li M, Wang Y, He X, Zheng N, Zhang J, et al. Improving AlphaFlow for efficient protein ensembles generation. In: International Conference on Machine Learning AI for Science workshop, Vienna, Austria; 2024.
Wang T, He X, Li M, Li Y, Bi R, Wang Y, et al. Ab initio characterization of protein molecular dynamics with AI2BMD. Nature 2024. https://doi.org/10.1038/s41586-024-08127-z.
DeVore K, Chiu PL. Probing structural perturbation of biomolecules by extracting Cryo-EM data heterogeneity. Biomolecules. 2022;12:628.
Zhao LH, Ma S, Sutkeviciute I, Shen DD, Zhou XE, de Waal PW, et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science. 2019;364:148–53.
Roy R, Al-Hashimi HM. AlphaFold3 takes a step toward decoding molecular behavior and biological computation. Nat Struct Mol Biol. 2024;31:997–1000.
Ren F, Ding X, Zheng M, Korzinkin M, Cai X, Zhu W, et al. AlphaFold accelerates artificial intelligence powered drug discovery: efficient discovery of a novel CDK20 small molecule inhibitor. Chem Sci. 2023;14:1443–52.
Borkakoti N, Thornton JM. AlphaFold2 protein structure prediction: Implications for drug discovery. Curr Opin Struct Biol. 2023;78:102526.
Francoeur PG, Masuda T, Sunseri J, Jia A, Iovanisci RB, Snyder I, et al. Three-dimensional convolutional neural networks and a cross-docked data set for structure-based drug design. J Chem Inf Model. 2020;60:4200–15.
Corso G, Deng A, Fry B, Polizzi N, Barzilay R, Jaakkola T. Deep confident steps to new pockets: strategies for docking generalization. In: International Conference on Learning Representations. Vienna Austria; 2024.
Hayes T, Rao R, Akin H, Sofroniew NJ, Oktay D, Lin Z, et al. Simulating 500 million years of evolution with a language model. bioRxiv 2024. https://doi.org/10.1101/2024.07.01.600583.
Fang Y, Zhang Q, Zhang N, Chen Z, Zhuang X, Shao X, et al. Knowledge graph-enhanced molecular contrastive learning with functional prompt. Nat Mach Intell. 2023;5:542–53.
Acknowledgements
The project was supported by the National Key R&D Program of China (2022YFC2703105 to HEX); The National Natural Science Foundation of China (32130022, 82121005); CAS Strategic Priority Research Program (XDB37030103 to HEX); Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to HEX); Shanghai Municipal Science and Technology Major Project (HEX); the Lingang Laboratory, Grant No. LG-GG-202204-01 (HEX). The authors acknowledge the Beijing Super Cloud Center (BSCC) for providing HPC resources that have contributed to the research results reported within this paper. URL: http://www.blsc.cn/.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, Xh., Li, Jr., Shen, Sy. et al. AlphaFold3 versus experimental structures: assessment of the accuracy in ligand-bound G protein-coupled receptors. Acta Pharmacol Sin 46, 1111–1122 (2025). https://doi.org/10.1038/s41401-024-01429-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41401-024-01429-y
Keywords
This article is cited by
-
Benchmarking co-folding methods to predict the structures of covalent protein–ligand complexes
Acta Pharmacologica Sinica (2026)
-
The Human Omnibus of Targetable Pockets
Journal of Cheminformatics (2025)
-
Molecular mechanisms of potato (Solanum tuberosum L.) transcription factor StbZIP1 in regulating saline-alkaline stress response through enhanced antioxidant capacity
Chemical and Biological Technologies in Agriculture (2025)
-
An update for AlphaFold3 versus experimental structures: assessing the precision of small molecule binding in GPCRs
Acta Pharmacologica Sinica (2025)
-
Neuropsychopharmacology in the era of artificial intelligence and biomolecule prediction software
NPP—Digital Psychiatry and Neuroscience (2025)