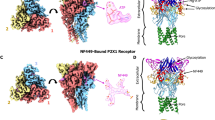
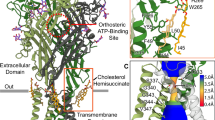
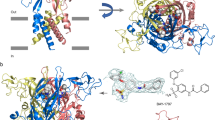
Abstract
As important modulators of human purinergic signaling, P2X1 receptors form homotrimers to transport calcium, regulating multiple physiological processes, and are long regarded as promising therapeutic targets for male contraception and inflammation. However, the development of drugs that target the P2X1 receptor, such as the antagonist NF449, is greatly hindered by the unclear molecular mechanism of ligand binding modes and receptor activation. Here, we report the structures of the P2X1 receptor in complex with the endogenous agonist ATP or the competitive antagonist NF449. The P2X1 receptor displays distinct conformational features when bound to different types of compounds. Despite coupling to the agonist ATP, the receptor adopts a desensitized conformation that arrests the ions in the transmembrane (TM) domain, aligning with the nature of the high desensitization rates of the P2X1 receptor within the P2X family. Interestingly, the antagonist NF449 not only occupies the orthosteric pocket of ATP but also interacts with the dorsal fin, left flipper, and head domains, suggesting a unique binding mode to perform both orthosteric and allosteric mechanisms of NF449 inhibition. Intriguingly, a novel lipid binding site adjacent to the TM helices and lower body of P2X1, which is critical for receptor activation, is identified. Further functional assay results and structural alignments reveal the high conservation of this lipid binding site in P2X receptors, indicating important modulatory roles upon lipid binding. Taken together, these findings greatly increase our understanding of the ligand binding modes and multiple modulatory mechanisms of the P2X1 receptor and shed light on the further development of P2X1-selective antagonists.
Keywords: Structural biology; Ligand binding mode; Ion channel; Purinergic P2X1 receptor
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The coordinates and cryo-EM density maps for the structures of the ATP-P2X1 and NF449-P2X1 complexes have been deposited in the PDB database with the identification codes 9LX5 and 9LXC, respectively, and in the Electron Microscopy Data Bank under accession codes EMD-63466 and EMD-63473, respectively.
References
Verkhratsky A, Burnstock G. Biology of purinergic signalling: its ancient evolutionary roots, its omnipresence and its multiple functional significance. Bioessays. 2014;36:660–70.
Lustig KD, Shiau AK, Brake AJ, Julius D. Expression cloning of an ATP receptor from mouse neuroblastoma cells. Proc Natl Acad Sci USA. 1993;90:5113–7.
Burnstock G. Purine and purinergic receptors. Brain Neurosci Adv. 2018;2:23.
North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–67.
Amobi NIB, Guillebaud J, Smith ICH. Perspective on the role of P2X-purinoceptor activation in human vas deferens contractility. Exp Physiol. 2012;97:583–602.
Bennetts FM, Mobbs JI, Ventura S, Thal DM. The P2X1 receptor as a therapeutic target. Purinergic Signal. 2022;18:421–33.
Burnstock G, Ralevic V. Purinergic signaling and blood vessels in health and disease. Pharmacol Rev. 2014;66:102–92.
Burnstock G. Pathophysiology and therapeutic potential of purinergic signaling. Pharmacol Rev. 2006;58:58–86.
Surprenant A, Schneider DA, Wilson HL, Galligan JJ, North RA. Functional properties of heteromeric P2X(1/5) receptors expressed in HEK cells and excitatory junction potentials in Guinea-pig submucosal arterioles. J Auton Nerv Syst. 2000;81:249–63.
Burnstock G. Physiology and pathophysiology of purinergic neurotransmission. Physiol Rev. 2007;87:659–797.
Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X4 ion channel in the closed state. Nature. 2009;460:592–8.
Hattori M, Gouaux E. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature. 2012;485:207–12.
McCarthy AE, Yoshioka C, Mansoor SE. Full-length P2X7 structures reveal how palmitoylation prevents channel desensitization. Cell. 2019;179:659–70.
Karasawa A, Kawate T. Structural basis for subtype-specific inhibition of the P2X7 receptor. eLife. 2016;5:e22153.
Kasuya G, Yamaura T, Ma XB, Nakamura R, Takemoto M, Nagumo H, et al. Structural insights into the competitive inhibition of the ATP-gated P2X receptor channel. Nat Commun. 2017;8:876.
Sheng D, Yue CX, Jin F, Wang Y, Ichikawa M, Yu Y, et al. Structural insights into the orthosteric inhibition of P2X receptors by non-ATP analog antagonists. eLife. 2024;12:RP92829.
Murrell-Lagnado RD. Regulation of P2X purinergic receptor signaling by cholesterol. Curr Top Membr. 2017;80:211–32.
White CW, Choong YT, Short JL, Exintaris B, Malone DT, Allen AM, et al. Male contraception via simultaneous knockout of α1A-adrenoceptors and P2X1-purinoceptors in mice. Proc Natl Acad Sci USA. 2013;110:20825–30.
Maitre B, Magnenat S, Heim V, Ravanat C, Evans RJ, De La Salle H, et al. The P2X1 receptor is required for neutrophil extravasation during lipopolysaccharide-induced lethal endotoxemia in mice. J Immunol. 2015;194:739–49.
Wéra O, Lecut C, Servais L, Hego A, Delierneux C, Jiang Z, et al. P2X1 ion channel deficiency causes massive bleeding in inflamed intestine and increases thrombosis. J Thromb Haemost. 2020;18:44–56.
Illes P, Müller CE, Jacobson KA, Grutter T, Nicke A, Fountain SJ, et al. Update of P2X receptor properties and their pharmacology: IUPHAR review 30. Br J Pharmacol. 2021;178:489–514.
Mulryan K, Gitterman DP, Lewis CJ, Vial C, Leckie BJ, Cobb AL, et al. Reduced vas deferens contraction and male infertility in mice lacking P2X1 receptors. Nature. 2000;403:86–89.
Rettinger J, Braun K, Hochmann H, Kassack MU, Ullmann H, Nickel P, et al. Profiling at recombinant homomeric and heteromeric rat P2X receptors identifies the suramin analogue NF449 as a highly potent P2X1 receptor antagonist. Neuropharmacology. 2005;48:461–8.
Brown SG, Kim YC, Kim SA, Jacobson KA, Burnstock G, King BF. Actions of a series of PPADS analogs at P2X1 and P2X3 receptors. Drug Dev Res. 2001;53:281–91.
Obrecht AS, Urban N, Schaefer M, Röse A, Kless A, Meents JE, et al. Identification of aurintricarboxylic acid as a potent allosteric antagonist of P2X1 and P2X3 receptors. Neuropharmacology. 2019;158:107749.
Tian M, Abdelrahman A, Baqi Y, Fuentes E, Azazna D, Spanier C, et al. Discovery and structure relationships of salicylanilide derivatives as potent, non-acidic P2X1 receptor antagonists. J Med Chem. 2020;63:6164–78.
Jacobson KA, Kim YC, Wildman SS, Mohanram A, Harden TK, Boyer JL, et al. A pyridoxine cyclic phosphate and its 6-azoaryl derivative selectively potentiate and antagonize activation of P2X1 receptors. J Med Chem. 1998;41:2201–6.
Kassack MU, Braun K, Ganso M, Ullmann H, Nickel P, Böing B, et al. Structure-activity relationships of analogues of NF449 confirm NF449 as the most potent and selective known P2X1 receptor antagonist. Eur J Med Chem. 2004;39:345–57.
Sattler C, Benndorf K. Enlightening activation gating in P2X receptors. Purinergic Signal. 2022;18:177–91.
Roberts JA, Evans RJ. Contribution of conserved polar glutamine, asparagine and threonine residues and glycosylation to agonist action at human P2X1 receptors for ATP. J Neurochem. 2006;96:843–52.
Bianchi BR, Lynch KJ, Touma E, Niforatos W, Burgard EC, Alexander KM, et al. Pharmacological characterization of recombinant human and rat P2X receptor subtypes. Eur J Pharmacol. 1999;376:127–38.
Kawate T. P2X receptor activation. Adv Exp Med Biol. 2017;1051:55–69.
Bernier LP, Ase AR, Séguéla P. Post-translational regulation of P2X receptor channels: modulation by phospholipids. Front Cell Neurosci. 2013;7:226.
Robinson LE, Shridar M, Smith P, Murrell-Lagnado RD. Plasma membrane cholesterol as a regulator of human and rodent P2X7 receptor activation and sensitization. J Biol Chem. 2014;289:31983–94.
Vial C, Evans RJ. Disruption of lipid rafts inhibits P2X1 receptor-mediated currents and arterial vasoconstriction. J Biol Chem. 2005;280:30705–11.
Wang J, Wang Y, Cui WW, Huang Y, Yang Y, Liu Y, et al. Druggable negative allosteric site of P2X3 receptors. Proc Natl Acad Sci USA. 2018;115:4939–44.
Shen C, Zhang Y, Cui W, Zhao Y, Sheng D, Teng X, et al. Structural insights into the allosteric inhibition of P2X4 receptors. Nat Commun. 2023;14:6437.
Allsopp RC, Evans RJ. The intracellular amino terminus plays a dominant role in desensitization of ATP-gated P2X receptor ion channels. J Biol Chem. 2011;286:44691–701.
Nicke A, Bäumert HG, Rettinger J, Eichele A, Lambrecht G, Mutschler E, et al. P2X1 and P2X3 receptors form stable trimers: a novel structural motif of ligand-gated ion channels. EMBO J. 1998;17:3016–28.
Torres GE, Egan TM, Voigt MM. Hetero-oligomeric assembly of P2X receptor subunits: specificities exist with regard to possible partners. J Biol Chem. 1999;274:6653–9.
Acknowledgements
The cryo-EM studies were performed at the electron microscopy facility of the Shanghai Institute of Materia Medica (SIMM), Chinese Academy of Sciences. We thank Yue Zhou from SIMM for cryo-EM data collection. This work was supported by the CAS Strategic Priority Research Program XDB37030100 (QZ).
Author information
Authors and Affiliations
Contributions
YTQ established and optimized the protein purification procedures, prepared protein samples for cryo-EM data collection, and helped with manuscript preparation. PPW performed the functional assays and helped with the data analysis. XL and YTC helped with plasmid cloning and protein preparation. LP expressed the proteins. LKZ helped with the protein purification and data analysis. ZBG planned and analyzed the experiments and helped with manuscript preparation. KC collected the cryo-EM data, performed the cryo-EM data processing and analysis, as well as model building and structure refinement, wrote the first version of the manuscript, and helped with the data analysis. QZ initiated the project, planned and analyzed the experiments, supervised the research, and finalized the manuscript with input from all coauthors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qiang, Yt., Wu, Pp., Liu, X. et al. Structural basis of the multiple ligand binding mechanisms of the P2X1 receptor. Acta Pharmacol Sin 46, 2564–2573 (2025). https://doi.org/10.1038/s41401-025-01512-y
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41401-025-01512-y