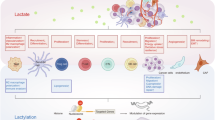
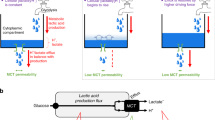
Abstract
Human monocarboxylate transporters (MCTs) are crucial for tumour cell glycolysis. Inhibiting MCT-mediated lactate transport can suppress the proliferation of solid tumours and enhance the efficacy of the immune system against tumours. Despite the importance of this transporter, the molecular mechanism of lactate transport by MCT1 remains elusive, hindering the development of targeted therapies. Here, we used principal component analysis to elucidate the allosteric mechanisms of the MCT family. Enhanced sampling revealed that specific residue pairs (E46-K289 and E376-R143) are essential for maintaining the inwards and outwards conformations of MCT1. Quantum chemical calculations and umbrella sampling demonstrated that lactate molecules and protons are co-transported sequentially, with K38 and R313 playing key roles in lactate translocation. On the basis of these data, we conducted a drug screening campaign targeting the core pocket of MCT1 and identified silybin as a selective MCT1 inhibitor. Silybin had significant inhibitory effects on tumour cells with high MCT1 expression. These findings provide a comprehensive understanding of the lactate transport mechanism of MCT1 and lay the groundwork for the rational design of antitumour drugs targeting MCT1.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–8.
Halestrap AP, Price NT. The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem J. 1999;343:281–99.
Bosshart PD, Kalbermatter D, Bonetti S, Fotiadis D. Mechanistic basis of L-lactate transport in the SLC16 solute carrier family. Nat Commun. 2019;10:2649.
Bosshart PD, Charles RP, Garibsingh RA, Schlessinger A, Fotiadis D. SLC16 family: from atomic structure to human disease. Trends Biochem Sci. 2021;46:28–40.
Meredith D, Christian HC. The SLC16 monocaboxylate transporter family. Xenobiotica. 2008;38:1072–106.
Felmlee MA, Jones RS, Rodriguez-Cruz V, Follman KE, Morris ME. Monocarboxylate transporters (SLC16): function, regulation, and role in health and disease. Pharmacol Rev. 2020;72:466–85.
Tosur M, Jeha GS. A novel intragenic SLC16A1 mutation associated with congenital hyperinsulinism. Glob Pediatr Health. 2017;4:2333794X17703462.
Wilson MC, Meredith D, Fox JE, Manoharan C, Davies AJ, Halestrap AP. Basigin (CD147) is the target for organomercurial inhibition of monocarboxylate transporter isoforms 1 and 4: the ancillary protein for the insensitive MCT2 is EMBIGIN (gp70). J Biol Chem. 2005;280:27213–21.
Walters DK, Arendt BK, Jelinek DF. CD147 regulates the expression of MCT1 and lactate export in multiple myeloma cells. Cell Cycle. 2013;12:3175–83.
Brooks GA. The science and translation of lactate shuttle theory. Cell Metab. 2018;27:757–85.
Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33.
Halestrap AP. Monocarboxylic acid transport. Compr Physiol. 2013;3:1611–43.
Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001.
Murray CM, Hutchinson R, Bantick JR, Belfield GP, Benjamin AD, Brazma D, et al. Monocarboxylate transporter MCT1 is a target for immunosuppression. Nat Chem Biol. 2005;1:371–6.
Draoui N, Schicke O, Fernandes A, Drozak X, Nahra F, Dumont A, et al. Synthesis and pharmacological evaluation of carboxycoumarins as a new antitumor treatment targeting lactate transport in cancer cells. Bioorg Med Chem. 2013;21:7107–17.
Ovens MJ, Davies AJ, Wilson MC, Murray CM, Halestrap AP. AR-C155858 is a potent inhibitor of monocarboxylate transporters MCT1 and MCT2 that binds to an intracellular site involving transmembrane helices 7-10. Biochem J. 2010;425:523–30.
Quanz M, Bender E, Kopitz C, Grünewald S, Schlicker A, Schwede W, et al. Preclinical efficacy of the novel monocarboxylate transporter 1 inhibitor BAY-8002 and associated markers of resistance. Mol Cancer Ther. 2018;17:2285–96.
Plummer R, Halford S, Jones P, Wedge S, Hirschberg S, Veal G, et al. A first-in-human first-in-class (FIC) trial of the monocarboxylate transporter 1 (MCT1) inhibitor AZD3965 in patients with advanced solid tumours. J Clin Oncol. 2017;35:2516–2516.
Xu B, Zhang M, Zhang B, Chi W, Ma X, Zhang W, et al. Embigin facilitates monocarboxylate transporter 1 localization to the plasma membrane and transition to a decoupling state. Cell Rep. 2022;40:111343.
Zhang B, Jin Q, Xu L, Li N, Meng Y, Chang S, et al. Cooperative transport mechanism of human monocarboxylate transporter 2. Nat Commun. 20205;11:2429.
Wang N, Jiang X, Zhang S, Zhu A, Yuan Y, Xu H, et al. Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates. Cell. 2021;184:370–83.
Brooks BR, Brooks CL 3rd, Mackerell AD Jr, Nilsson L, Petrella RJ, Roux B, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–614.
Case DA, Aktulga HM, Belfon K, Ben-Shalom IY, Berryman JT, Brozell SR, et al. Amber 2022. San Francisco: University of California; 2022.
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 16. 2016: Wallingford, CT.
Lu T, Chen F. Multiwfn: a multifunctional wavefunction analyzer. J Comput Chem. 2012;33:580–92.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61.
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91.
Arce-Molina R, Cortés-Molina F, Sandoval PY, Galaz A, Alegría K, Schirmeier S, et al. A highly responsive pyruvate sensor reveals pathway-regulatory role of the mitochondrial pyruvate carrier MPC. Elife. 2020;9:e53917.
Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–86.
Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8.
Zhang Y, Doruker P, Kaynak B, Zhang S, Krieger J, Li H, et al. Intrinsic dynamics is evolutionarily optimized to enable allosteric behavior. Curr Opin Struct Biol. 2020;62:14–21.
Bakan A, Meireles LM, Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27:1575–7.
Bahar I, Lezon TR, Yang LW, Eyal E. Global dynamics of proteins: bridging between structure and function. Annu Rev Biophys. 2010;39:23–42.
Pinheiro C, Longatto-Filho A, Azevedo-Silva J, Casal M, Schmitt FC, Baltazar F. Role of monocarboxylate transporters in human cancers: state of the art. J Bioenerg Biomembr. 2012;44:127–39.
Halestrap AP. The SLC16 gene family - structure, role and regulation in health and disease. Mol Aspects Med. 2013;34:337–49.
Polański R, Hodgkinson CL, Fusi A, Nonaka D, Priest L, Kelly P, et al. Activity of the monocarboxylate transporter 1 inhibitor AZD3965 in small cell lung cancer. Clin Cancer Res. 2014;20:926–37.
Smirnova IN, Kasho V, Kaback HR. Protonation and sugar binding to LacY. Proc Natl Acad Sci USA. 2008;105:8896–901.
Loguercio C, Andreone P, Brisc C, Brisc MC, Bugianesi E, Chiaramonte M, et al. Silybin combined with phosphatidylcholine and vitamin E in patients with nonalcoholic fatty liver disease: a randomized controlled trial. Free Radic Biol Med. 2012;52:1658–65.
Flaig TW, Gustafson DL, Su LJ, Zirrolli JA, Crighton F, Harrison GS, et al. A phase I and pharmacokinetic study of silybin-phytosome in prostate cancer patients. Invest New Drugs. 2007;25:139–46.
Cheung CW, Gibbons N, Johnson DW, Nicol DL. Silibinin–a promising new treatment for cancer. Anticancer Agents Med Chem. 2010;10:186–95.
Javed S, Kohli K, Ali M. Reassessing bioavailability of silymarin. Altern Med Rev. 2011;16:239–49.
Wang Y, Zhang L, Wang Q, Zhang D. Recent advances in the nanotechnology-based drug delivery of silybin. J Biomed Nanotechnol. 2014;10:543–58.
Waks AG, Winer EP. Breast cancer treatment a review. JAMA. 2019;321:288–300.
Jin MZ, Jin WL. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct Target Ther. 2020;5:166.
Shi S, Ma B, Ji Q, Guo S, An H, Ye S. Identification of a druggable pocket of the calcium-activated chloride channel TMEM16A in its open state. J Biol Chem. 2023;299:104780.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 82404583 to SS). Hebei Province Yanzhao Golden Platform Talent Gathering Plan Backbone Talent Project (Education Platform) (Grant No. HJYB202524 to SS). Ministry of Science and Technology (2024YFA0916800 to BHX, 2020YFA0908500 to SY, 2024YFC3407300 to YXW). The Independent Innovation Fund of Tianjin University (2023XQM-0050 to BHX). We thank the Haihe Laboratory of Sustainable Chemical Transformations for financial support. This research was supported by the Medical Science Data Center of Hebei Medical University.
Author information
Authors and Affiliations
Contributions
SS: conceptualization, methodology, writing - original draft. JCL: investigation, validation. XYZ: data curation. ZLL: methodology. YXW: formal analysis, supervision. BHX: resources, writing - original draft. SY: project administration, writing - original draft.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, S., Li, Jc., Zhou, Xy. et al. Transport mechanism and drug discovery of human monocarboxylate transporter 1. Acta Pharmacol Sin 46, 2323–2333 (2025). https://doi.org/10.1038/s41401-025-01517-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41401-025-01517-7