Abstract
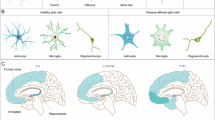
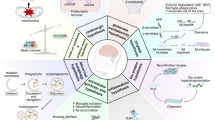
Neurodegenerative diseases (NDDs) are characterized by progressive neuronal dysfunction and anatomical changes caused by neuron loss and gliosis, ultimately leading to severe declines in brain function. While these disorders arise from a variety of pathological mechanisms, a common molecular feature is the accumulation of misfolded proteins, which occurs both inside and outside neurons. For example, Alzheimer’s disease (AD) is defined by extracellular β-amyloid plaques and intracellular tau neurofibrillary tangles. These pathological protein aggregates are often resistant to traditional small molecule drugs. Recent advances in proximity-inducing chimeras such as proteolysis-targeting chimeras (PROTACs), lysosome-targeting chimeras (LYTACs), autophagy-targeted chimeras (AUTOTACs), dephosphorylation-targeting chimeras (DEPTACs) and ribonuclease-targeting chimeras (RIBOTACs) offer promising strategies to eliminate pathological proteins or mRNAs through intracellular degradation pathways. These innovative approaches open avenues for developing new therapies for NDDs. In this review we summarize the regulatory mechanisms of protein aggregation, highlight the advancements in proximity-inducing modalities for NDDs, and discuss the current challenges and future directions in therapeutic development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Wilson DM 3rd, Cookson MR, Van Den Bosch L, Zetterberg H, Holtzman DM, Dewachter I. Hallmarks of neurodegenerative diseases. Cell. 2023;186:693–714.
Temple S. Advancing cell therapy for neurodegenerative diseases. Cell Stem Cell. 2023;30:512–29.
Wendimu MY, Hooks SB. Microglia phenotypes in aging and neurodegenerative diseases. Cells. 2022;11:2091–143.
Guo J, Huang X, Dou L, Yan M, Shen T, Tang W, et al. Aging and aging-related diseases: from molecular mechanisms to interventions and treatments. Signal Transduct Target Ther. 2022;7:391.
Békés M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 2022;21:181–200.
Ahn G, Riley NM, Kamber RA, Wisnovsky S, Moncayo von Hase S, Bassik MC, et al. Elucidating the cellular determinants of targeted membrane protein degradation by lysosome-targeting chimeras. Science. 2023;382:eadf6249.
Li YY, Yang Y, Zhang RS, Ge RX, Xie SB. Targeted degradation of membrane and extracellular proteins with LYTACs. Acta Pharmacol Sin. 2025;46:1–7.
Ji CH, Kim HY, Lee MJ, Heo AJ, Park DY, Lim S, et al. The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system. Nat Commun. 2022;13:904.
Tan S, Wang D, Fu Y, Zheng H, Liu Y, Lu B. Targeted clearance of mitochondria by an autophagy-tethering compound (ATTEC) and its potential therapeutic effects. Sci Bull. 2023;68:3013–26.
Su JF, Xiao Y, Wei LY, Lei HY, Sun F, Wang WX, et al. A new tau dephosphorylation-targeting chimera for the treatment of tauopathies. Acta Pharmacol Sin. 2024;45:2267–76.
Bonet-Aleta J, Maehara T, Craig BA, Bernardes GJL. Small molecule RNA degraders. Angew Chem Int Ed Engl. 2024;63:e202412925.
Kiss L, Rhinesmith T, Luptak J, Dickson CF, Weidenhausen J, Smyly S, et al. Trim-Away ubiquitinates and degrades lysine-less and N-terminally acetylated substrates. Nat Commun. 2023;14:2160.
Zhu W, Zhang W, Chen J, Tong Y, Xu F, Pang J. Discovery of effective dual PROTAC degraders for neurodegenerative disease-associated aggregates. J Med Chem. 2024;67:3448–66.
Fang Y, Wang J, Zhao M, Zheng Q, Ren C, Wang Y, et al. Progress and challenges in targeted protein degradation for neurodegenerative disease therapy. J Med Chem. 2022;65:11454–77.
Zhao L, Zhao J, Zhong K, Tong A, Jia D. Targeted protein degradation: mechanisms, strategies and application. Signal Transduct Target Ther. 2022;7:113.
Singh J, Habean ML, Panicker N. Inflammasome assembly in neurodegenerative diseases. Trends Neurosci. 2023;46:814–31.
Hung ST, Linares GR, Chang WH, Eoh Y, Krishnan G, Mendonca S, et al. PIKFYVE inhibition mitigates disease in models of diverse forms of ALS. Cell. 2023;186:786–802.
Chen Y, Song S, Parhizkar S, Lord J, Zhu Y, Strickland MR, et al. APOE3ch alters microglial response and suppresses Aβ-induced tau seeding and spread. Cell. 2024;187:428–45.
Ossenkoppele R, van der Kant R, Hansson O. Tau biomarkers in Alzheimer’s disease: towards implementation in clinical practice and trials. Lancet Neurol. 2022;21:726–34.
Li Q, Pan W, Zhou J, Yu H, Xie S. Targeting protein aggregation for the treatment of neurodegenerative diseases. Med Plus. 2024;1:100005.
Morris HR, Spillantini MG, Sue CM, Williams-Gray CH. The pathogenesis of Parkinson’s disease. Lancet. 2024;403:293–304.
Tabrizi SJ, Estevez-Fraga C, van Roon-Mom WMC, Flower MD, Scahill RI, Wild EJ, et al. Potential disease-modifying therapies for Huntington’s disease: lessons learned and future opportunities. Lancet Neurol. 2022;21:645–58.
Jucker M, Walker LC. Alzheimer’s disease: from immunotherapy to immunoprevention. Cell. 2023;186:4260–70.
Knezevic E, Nenic K, Milanovic V, Knezevic NN. The role of cortisol in chronic stress, neurodegenerative diseases, and psychological disorders. Cells. 2023;12:2726.
Parra Bravo C, Naguib SA, Gan L. Cellular and pathological functions of tau. Nat Rev Mol Cell Biol. 2024;25:845–64.
Duan P, Dregni AJ, Mammeri NE, Hong M. Structure of the nonhelical filament of the Alzheimer’s disease tau core. Proc Natl Acad Sci USA. 2023;120:e2310067120.
Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, Bondulich MK, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci USA. 2013;110:2366–70.
Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–91.
Aldous SG, Smith EJ, Landles C, Osborne GF, Cañibano-Pico M, Nita IM, et al. A CAG repeat threshold for therapeutics targeting somatic instability in Huntington’s disease. Brain. 2024;147:1784–98.
Aviner R, Lee TT, Masto VB, Li KH, Andino R, Frydman J. Polyglutamine-mediated ribotoxicity disrupts proteostasis and stress responses in Huntington’s disease. Nat Cell Biol. 2024;26:892–902.
Harel I, Chen YR, Ziv I, Singh PP, Heinzer D, Navarro Negredo P, et al. Identification of protein aggregates in the aging vertebrate brain with prion-like and phase-separation properties. Cell Rep. 2024;43:112787.
Lu M, Williamson N, Mishra A, Michel CH, Kaminski CF, Tunnacliffe A, et al. Structural progression of amyloid-β Arctic mutant aggregation in cells revealed by multiparametric imaging. J Biol Chem. 2019;294:1478–87.
Goutman SA, Hardiman O, Al-Chalabi A, Chió A, Savelieff MG, Kiernan MC, et al. Recent advances in the diagnosis and prognosis of amyotrophic lateral sclerosis. Lancet Neurol. 2022;21:480–93.
Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA. 2001;98:8554–9.
Yang Z, Pang Q, Zhou J, Xuan C, Xie S. Leveraging aptamers for targeted protein degradation. Trends Pharmacol Sci. 2023;44:776–85.
Chen M, Zhou P, Kong Y, Li J, Li Y, Zhang Y, et al. Inducible degradation of oncogenic nucleolin using an aptamer-based PROTAC. J Med Chem. 2023;66:1339–48.
Kong L, Meng F, Zhou P, Ge R, Geng X, Yang Z, et al. An engineered DNA aptamer-based PROTAC for precise therapy of p53-R175H hotspot mutant-driven cancer. Sci Bull. 2024;69:2122–35.
Chu TT, Gao N, Li QQ, Chen PG, Yang XF, Chen YX, et al. Specific Knockdown of endogenous Tau protein by peptide-directed ubiquitin-proteasome degradation. Cell Chem Biol. 2016;23:453–61.
Lu M, Liu T, Jiao Q, Ji J, Tao M, Liu Y, et al. Discovery of a Keap1-dependent peptide PROTAC to knockdown Tau by ubiquitination-proteasome degradation pathway. Eur J Med Chem. 2018;146:251–9.
Silva MC, Nandi G, Donovan KA, Cai Q, Berry BC, Nowak RP, et al. Discovery and optimization of Tau targeted protein degraders enabled by patient induced pluripotent stem cells-derived neuronal models of tauopathy. Front Cell Neurosci. 2022;16:801179.
Wang W, Zhou Q, Jiang T, Li S, Ye J, Zheng J, et al. A novel small-molecule PROTAC selectively promotes tau clearance to improve cognitive functions in Alzheimer-like models. Theranostics. 2021;11:5279–95.
Qu J, Ren X, Xue F, He Y, Zhang R, Zheng Y, et al. Specific knockdown of α-synuclein by peptide-directed proteasome degradation rescued its associated neurotoxicity. Cell Chem Biol. 2020;27:763.
Tong Y, Zhu W, Chen J, Wen T, Xu F, Pang J. Discovery of small-molecule degraders for alpha-synuclein aggregates. J Med Chem. 2023;66:7926–42.
Tomoshige S, Nomura S, Ohgane K, Hashimoto Y, Ishikawa M. Discovery of small molecules that induce the degradation of Huntingtin. Angew Chem Int Ed Engl. 2017;56:11530–3.
Tomoshige S, Nomura S, Ohgane K, Hashimoto Y, Ishikawa M. Degradation of huntingtin mediated by a hybrid molecule composed of IAP antagonist linked to phenyldiazenyl benzothiazole derivative. Bioorg Med Chem Lett. 2018;28:707–10.
Feldman EL, Goutman SA, Petri S, Mazzini L, Savelieff MG, Shaw PJ, et al. Amyotrophic lateral sclerosis. Lancet. 2022;400:1363–80.
Piol D, Robberechts T, Da Cruz S. Lost in local translation: TDP-43 and FUS in axonal/neuromuscular junction maintenance and dysregulation in amyotrophic lateral sclerosis. Neuron. 2023;111:1355–80.
Tseng YL, Lu PC, Lee CC, He RY, Huang YA, Tseng YC, et al. Degradation of neurodegenerative disease-associated TDP-43 aggregates and oligomers via a proteolysis-targeting chimera. J Biomed Sci. 2023;30:27.
Gao N, Chu TT, Li QQ, Lim YJ, Qiu T, Ma MR, et al. Hydrophobic tagging-mediated degradation of Alzheimer’s disease related Tau. RSC Adv. 2017;7:40362–6.
Gao N, Huang YP, Chu TT, Li QQ, Zhou B, Chen YX, et al. TDP-43 specific reduction induced by Di-hydrophobic tags conjugated peptides. Bioorg Chem. 2019;84:254–9.
Jeong J, Usman M, Li Y, Zhou XZ, Lu KP. Pin1-catalyzed conformation changes regulate protein ubiquitination and degradation. Cells. 2024;13:731.
Holwek E, Opinc-Rosiak A, Sarnik J, Makowska J. Ro52/TRIM21 - From host defense to autoimmunity. Cell Immunol. 2023;393-394:104776.
Mevissen TET, Prasad AV, Walter JC. TRIM21-dependent target protein ubiquitination mediates cell-free Trim-Away. Cell Rep. 2023;42:112125.
Xie S, Zhang L, Dong D, Ge R, He Q, Fan C, et al. HDAC6 regulates antibody-dependent intracellular neutralization of viruses via deacetylation of TRIM21. J Biol Chem. 2020;295:14343–51.
Benn J, Cheng S, Keeling S, Smith AE, Vaysburd MJ, Böken D, et al. Aggregate-selective removal of pathological tau by clustering-activated degraders. Science. 2024;385:1009–16.
Miller LVC, Papa G, Vaysburd M, Cheng S, Sweeney PW, Smith A, et al. Co-opting templated aggregation to degrade pathogenic tau assemblies and improve motor function. Cell. 2024;187:5967–80.
Dewey JA, Delalande C, Azizi SA, Lu V, Antonopoulos D, Babnigg G. Molecular glue discovery: current and future approaches. J Med Chem. 2023;66:9278–96.
Lu P, Cheng Y, Xue L, Ren X, Xu X, Chen C, et al. Selective degradation of multimeric proteins by TRIM21-based molecular glue and PROTAC degraders. Cell. 2024;187:7126–42.
Nixon RA, Rubinsztein DC. Mechanisms of autophagy-lysosome dysfunction in neurodegenerative diseases. Nat Rev Mol Cell Biol. 2024;25:926–46.
Fan X, Jin WY, Lu J, Wang J, Wang YT. Rapid and reversible knockdown of endogenous proteins by peptide-directed lysosomal degradation. Nat Neurosci. 2014;17:471–80.
Bauer PO, Goswami A, Wong HK, Okuno M, Kurosawa M, Yamada M, et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28:256–63.
Lee J, Sung KW, Bae EJ, Yoon D, Kim D, Lee JS, et al. Targeted degradation of α-synuclein aggregates in Parkinson’s disease using the AUTOTAC technology. Mol Neurodegener. 2023;18:41.
Mei L, Chen X, Wei F, Huang X, Liu L, Yao J, et al. Tethering ATG16L1 or LC3 induces targeted autophagic degradation of protein aggregates and mitochondria. Autophagy. 2023;19:2997–3013.
Li Z, Wang C, Wang Z, Zhu C, Li J, Sha T, et al. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature. 2019;575:203–9.
He H, Zhou C, Chen X. ATNC: versatile nanobody chimeras for autophagic degradation of intracellular unligandable and undruggable proteins. J Am Chem Soc. 2023;145:24785–95.
Zhang D, Duque-Jimenez J, Facchinetti F, Brixi G, Rhee K, Feng WW, et al. Transferrin receptor targeting chimeras for membrane protein degradation. Nature. 2025;638:787–95.
Huang B, Abedi M, Ahn G, Coventry B, Sappington I, Tang C, et al. Designed endocytosis-inducing proteins degrade targets and amplify signals. Nature. 2025;638:796–804.
Zhang S, Zhu R, Pan B, Xu H, Olufemi MF, Gathagan RJ, et al. Post-translational modifications of soluble α-synuclein regulate the amplification of pathological α-synuclein. Nat Neurosci. 2023;26:213–25.
Wesseling H, Mair W, Kumar M, Schlaffner CN, Tang S, Beerepoot P, et al. Tau PTM profiles identify patient heterogeneity and stages of Alzheimer’s disease. Cell. 2020;183:1699–713.
Zheng J, Tian N, Liu F, Zhang Y, Su J, Gao Y, et al. A novel dephosphorylation targeting chimera selectively promoting tau removal in tauopathies. Signal Transduct Target Ther. 2021;6:269.
Hu Z, Chen PH, Li W, Douglas T, Hines J, Liu Y, et al. Targeted Dephosphorylation of Tau by Phosphorylation Targeting Chimeras (PhosTACs) as a therapeutic modality. J Am Chem Soc. 2023;145:4045–55.
Su J, Xiao Y, Wei L, Lei H, Sun F, Wang W, et al. Generation of tau dephosphorylation-targeting chimeras for the treatment of Alzheimer’s disease and related tauopathies. Sci Bull. 2024;69:1137–52.
Tong Y, Zhang P, Yang X, Liu X, Zhang J, Grudniewska M, et al. Decreasing the intrinsically disordered protein α-synuclein levels by targeting its structured mRNA with a ribonuclease-targeting chimera. Proc Natl Acad Sci USA. 2024;121:e2306682120.
Bush JA, Aikawa H, Fuerst R, Li Y, Ursu A, Meyer SM, et al. Ribonuclease recruitment using a small molecule reduced c9ALS/FTD r(G(4)C(2)) repeat expansion in vitro and in vivo ALS models. Sci Transl Med. 2021;13:eabd5991.
Li Y, Song J, Zhou P, Zhou J, Xie S. Targeting undruggable transcription factors with PROTACs: advances and perspectives. J Med Chem. 2022;65:10183–94.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32270892 and 32200613) and the Shandong Provincial Natural Science Foundation (ZR2021MC157).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ge, Rx., Chen, M., Li, Qc. et al. Targeting neurodegenerative disease-associated protein aggregation with proximity-inducing modalities. Acta Pharmacol Sin 46, 2337–2346 (2025). https://doi.org/10.1038/s41401-025-01538-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41401-025-01538-2
Keywords
This article is cited by
-
DNA nanoflower Oligo-PROTAC for targeted degradation of FUS to treat neurodegenerative diseases
Nature Communications (2025)