Abstract
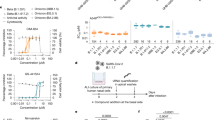
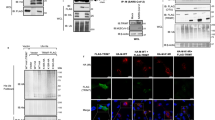
We previously reported that carrimycin could inhibit pan-coronavirus including HCoV-229E, HCoV-OC43 and SARS-CoV-2. We found that carrimycin targeted the post-entry replicative events in coronavirus infection. Carrimycin could impede the viral protein translation switch from ORF1a to ORF1b by targeting programmed -1 ribosomal frameshifting (-1PRF). Carrimycin could also inhibit the newly synthesized (nascent) viral RNA. In this study we investigated whether carrimycin also inhibited the newly emerged SARS-CoV-2 variants. We showed that carrimycin (1.25–10 µM) dose-dependently inhibited both viral RNA and protein levels in Vero E6 cells. We further demonstrated that carrimycin disrupted the formation of SARS-CoV-2 double membrane vesicles (DMVs), and identified the host transmembrane protein B (TMEM41B) as the key factor involved in this process. Overexpression of TMEM41B increased viral protein levels and mRNA levels, whereas TMEM41B knockdown reduced viral replication including HCoV-229E, HCoV-OC43 and SARS-CoV-2. Moreover, overexpression of TMEM41B partially reversed the inhibitory effect of carrimycin, suggesting that carrimycin indeed exerted antiviral effects through regulation of TMEM41B. We revealed that carrimycin directly bound to TMEM41B and induced its K48 ubiquitination degradation, thereby inhibiting viral replication. These results expand the understanding of carrimycin’s antiviral mechanisms, particularly its antiviral activity, and enrich our knowledge about the role of host factors in regulating viral replication.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Holmes KV, Lai M. Coronaviridae: the viruses and their replication. Fields Virol. 1996;1:1075–93.
Lamers MM, Haagmans BL. SARS-CoV-2 pathogenesis. Nat Rev Microbiol. 2022;20:270–84.
Cobar O, Cobar S. Omicron Variants World Prevalence, 174 WHO COVID-19 Epidemiological Update, ECDC Communicable Disease Threat Report, and CDC COVID Data Tracker Review. 2024;12:1–57.
Chan JF, Yuan S, Chu H, Sridhar S, Yuen KY. COVID-19 drug discovery and treatment options. Nat Rev Microbiol. 2024;22:391–407.
He W, Yang C, Zhao X, Wang Y. Antimicrobial activity of bitespiramycin, a new genetically engineered macrolide. Bioorg Med Chem Lett. 2017;27:4576–7.
Vázquez-Laslop N, Mankin AS. How macrolide antibiotics work. Trends Biochem Sci. 2018;43:668–84.
Yan H, Sun J, Wang K, Wang H, Wu S, Bao L, et al. Repurposing carrimycin as an antiviral agent against human coronaviruses, including the currently pandemic SARS-CoV-2. Acta Pharm Sin B. 2021;11:2850–58.
Li H, Li J, Li J, Li H, Wang X, Jiang J, et al. Carrimycin inhibits coronavirus replication by decreasing the efficiency of programmed–1 ribosomal frameshifting through directly binding to the RNA pseudoknot of viral frameshift-stimulatory element. Acta Pharm Sin B. 2024;14:2567–80.
Snijder EJ, Limpens RW, de Wilde AH, de Jong AWM, Zevenhoven-Dobbe JC, Maier HJ, et al. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715.
Huang Y, Wang T, Zhong L, Zhang W, Zhang Y, Yu X, et al. Molecular architecture of coronavirus double-membrane vesicle pore complex. Nature. 2024;633:224–31.
Wolff G, Melia CE, Snijder EJ, Bárcena M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020;28:1022–33.
Yan L, Zhang Y, Ge J, Zheng L, Gao Y, Wang T, et al. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat Commun. 2020;11:5874.
Ahn DG, Choi JK, Taylor DR, Oh JW. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch Virol. 2012;157:2095–104.
Iketani S, Hong SJ, Sheng J, Bahari F, Culbertson B, Atanaki FF, et al. Functional map of SARS-CoV-2 3CL protease reveals tolerant and immutable sites. Cell Host Microbe. 2022;30:1354–62.e6.
Schneider WM, Luna JM, Hoffmann HH, Sánchez-Rivera FJ, Leal AA, Ashbrook AW, et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184:120–32.e14.
Sun L, Zhao C, Fu Z, Fu Y, Su Z, Li Y, et al. Genome-scale CRISPR screen identifies TMEM41B as a multi-function host factor required for coronavirus replication. PLoS Pathog. 2021;17:e1010113.
Ji M, Li M, Sun L, Zhao H, Li Y, Zhou L, et al. VMP1 and TMEM41B are essential for DMV formation during β-coronavirus infection. J Cell Biol. 2022;221:e202112081.
Trimarco JD, Heaton BE, Chaparian RR, Burke KN, Binder RA, Gray GC, et al. TMEM41B is a host factor required for the replication of diverse coronaviruses including SARS-CoV-2. PLoS Pathog. 2021;17:e1009599.
Huang F, Zeng X, Kim W, Balasubramani M, Fortian A, Gygi SP, et al. Lysine 63-linked polyubiquitination is required for EGF receptor degradation. Proc Natl Acad Sci USA. 2013;110:15722–7.
Steiner S, Kratzel A, Barut GT, Lang RM, Aguiar Moreira E, Thomann L, et al. SARS-CoV-2 biology and host interactions. Nat Rev Microbiol. 2024;22:206–25.
Hoffmann HH, Schneider WM, Rozen-Gagnon K, Miles LA, Schuster F, Razooky B, et al. TMEM41B is a pan-flavivirus host factor. Cell. 2021;184:133–48.e20.
Yousefi M, Lee WS, Yan B, Cui L, Yong CL, Yap X, et al. TMEM41B and VMP1 modulate cellular lipid and energy metabolism for facilitating dengue virus infection. PLoS Pathog. 2022;18:e1010763.
Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, Van Der Meer Y, Koster AJ, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226.
Blanchard E, Roingeard P. Virus‐induced double‐membrane vesicles. Cell Microbiol. 2015;17:45–50.
Acknowledgements
We gratefully acknowledge Dr. Jing-dong Song (National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention) for technical supports and advice on the Transmission microscopy analysis. This work was financially supported by National Natural Science Foundation of China (82151525, 82003818, 82394464) and CAMS Innovation Fund for Medical Sciences (2021-I2M-1-030, 2022-I2M-CoV19-001), China.
Author information
Authors and Affiliations
Contributions
YHL, HQW and KW designed the experiments. YHL and JDJ directed the study. KW, HQW, GY, SW, HYY and MYW contributed to executing the described experiments. KW and HQW were responsible for the analysis of the data and preparation the manuscript. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, K., Wang, Hq., Yang, G. et al. Carrimycin exhibited broad spectrum inhibitory activities against coronaviruses replication through down-regulating host factor TMEM41B. Acta Pharmacol Sin 46, 2006–2015 (2025). https://doi.org/10.1038/s41401-025-01577-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41401-025-01577-9