Abstract
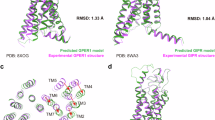
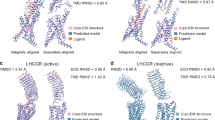
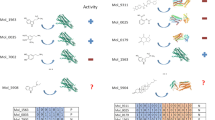
G protein-coupled receptors (GPCRs) are key drug discovery targets with many of them modulated by small molecules via diverse binding mechanisms. AlphaFold3, a leading structure prediction tool, models GPCR-small molecule complexes, but its accuracy remains insufficiently evaluated. In this study we compared 74 AlphaFold3-predicted structures to experimental counterparts. We showed that while AlphaFold3 accurately captured global receptor architecture and orthosteric binding pockets, which was consistent with our previous research, its ligand positioning was highly variable and often inaccurate, rendering predictions unreliable, particularly for allosteric modulators. The significant divergence from experimental structures, particularly for complex ligand interactions, highlighted AlphaFold3’s limitations and underscored that experimental structures remained essential for validating ligand-binding accuracy in GPCR complexes. These findings suggest that while AlphaFold3 offers potential for structure-based drug design, its current inaccuracies necessitate substantial refinement and integration with experimental data. This study highlights the limitation of AlphaFold3 in predicting small molecule binding and reinforces the critical role of high-resolution experimental validation for reliable GPCR-ligand interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
23 July 2025
In this article the acknowledgment section has been omitted. This has been corrected.
References
Albanese KI, Barbe S, Tagami S, Woolfson DN, Schiex T. Computational protein design. Nat Rev Methods Prim. 2025;5:13.
Callaway E. Chemistry Nobel goes to developers of AlphaFold AI that predicts protein structures. Nature. 2024;634:525–6.
Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500.
Beck H, Härter M, Haß B, Schmeck C, Baerfacker L. Small molecules and their impact in drug discovery: a perspective on the occasion of the 125th anniversary of the bayer chemical research laboratory. Drug Discov Today. 2022;27:1560–74.
Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–42.
Zhao L, He X, Jiang H, Cheng X. Computational characterization of transducer recognition of β2 adrenergic receptor. Biochem Biophys Res Commun. 2022;592:67–73.
Lu S, He X, Yang Z, Chai Z, Zhou S, Wang J, et al. Activation pathway of a G protein-coupled receptor uncovers conformational intermediates as targets for allosteric drug design. Nat Commun. 2021;12:4721.
Anantakrishnan S, Naganathan AN. Thermodynamic architecture and conformational plasticity of GPCRs. Nat Commun. 2023;14:128.
Zhou XE, Melcher K, Xu HE. Structural biology of G protein-coupled receptor signaling complexes. Protein Sci. 2019;28:487–501.
Cong X, Ren W, Pacalon J, Xu R, Xu L, Li X, et al. Large-scale G protein-coupled olfactory receptor–ligand pairing. ACS Cent Sci. 2022;8:379–87.
Fan W, Xu Y, He X, Luo P, Zhu J, Li J, et al. Molecular basis for the activation of PAF receptor by PAF. Cell Rep. 2024;43:114422.
Duan J, He X, Li S, Xu HE. Cryo-electron microscopy for GPCR research and drug discovery in endocrinology and metabolism. Nat Rev Endocrinol. 2024;20:349–65.
He X, Li J, Shen S, Xu HE. AlphaFold3 versus experimental structures: assessment of the accuracy in ligand-bound G protein-coupled receptors. Acta Pharmacol Sin. 2024;46:1111–22.
Zhuang Y, Wang Y, He B, He X, Zhou XE, Guo S, et al. Molecular recognition of morphine and fentanyl by the human μ-opioid receptor. Cell. 2022;185:4361–75.
Lee Y, Hou X, Lee JH, Nayak A, Alexander V, Sharma PK, et al. Subtle chemical changes cross the boundary between agonist and antagonist: new A3 adenosine receptor homology models and structural network analysis can predict this boundary. J Med Chem. 2021;64:12525–36.
Liu H, Zhang Q, He X, Jiang M, Wang S, Yan X, et al. Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84. Nat Commun. 2023;14:3271.
Maiti S, Singh A, Maji T, Saibo NV, De S. Experimental methods to study the structure and dynamics of intrinsically disordered regions in proteins. Curr Res Struct Biol. 2024;7:100138.
Costa MGS, Gur M, Krieger JM, Bahar I. Computational biophysics meets cryo-EM revolution in the search for the functional dynamics of biomolecular systems. Wiley Interdiscip Rev Comput Mol Sci. 2024;14:e1689.
Li X, Shen C, Zhu H, Yang Y, Wang Q, Yang J, et al. A high-quality data set of protein–ligand binding interactions via comparative complex structure modeling. J Chem Inf Model. 2024;64:2454–66.
Lan J, He X, Ren Y, Wang Z, Zhou H, Fan S, et al. Structural insights into the SARS-CoV-2 omicron RBD-ACE2 interaction. Cell Res. 2022;32:593–5.
Liu H, Zheng Y, Wang Y, Wang Y, He X, Xu P, et al. Recognition of methamphetamine and other amines by trace amine receptor TAAR1. Nature. 2023;624:663–71.
Ciancetta A, Jacobson KA. Breakthrough in GPCR crystallography and its impact on computer-aided drug design. Methods Mol Biol. 2018;1705:45–72.
Duan J, Liu H, Zhao F, Yuan Q, Ji Y, Cai X, et al. GPCR activation and GRK2 assembly by a biased intracellular agonist. Nature. 2023;620:676–81.
He X, You C, Jiang H, Jiang Y, Xu HE, Cheng X. AlphaFold2 versus experimental structures: evaluation on G protein-coupled receptors. Acta Pharmacol Sin. 2023;44:1–7.
Meli R, Biggin PC. spyrmsd: symmetry-corrected RMSD calculations in Python. J Cheminform. 2020;12:49.
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49.
Rimac H, Grishina M, Potemkin V. Use of the complementarity principle in docking procedures: a new approach for evaluating the correctness of binding poses. J Chem Inf Model. 2021;61:1801–13.
Ibrahim P, Wifling D, Clark T. Universal activation index for class A GPCRs. J Chem Inf Model. 2019;59:3938–45.
Zhao L, He Q, Yuan Q, Gu Y, He X, Shan H, et al. Conserved class B GPCR activation by a biased intracellular agonist. Nature. 2023;621:635–41.
Otanuly M, Kubitschke M, Masseck OA. A bright future? A perspective on class C GPCR based genetically encoded biosensors. ACS Chem Neurosci. 2024;15:889–97.
He X, Ni D, Lu S, Zhang J. Characteristics of allosteric proteins, sites, and modulators. Adv Exp Med Biol. 2019;1163:107–39.
Zhang M, Chen T, Lu X, Lan X, Chen Z, Lu S. G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery. Signal Transduct Target Ther. 2024;9:88.
Kruse AC, Ring AM, Manglik A, Hu J, Hu K, Eitel K, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–6.
Lu S, He X, Ni D, Zhang J. Allosteric modulator discovery: from serendipity to structure-based design. J Med Chem. 2019;62:6405–21.
He X, Duan J, Ji Y, Zhao L, Jiang H, Jiang Y, et al. Hinge region mediates signal transmission of luteinizing hormone and chorionic gonadotropin receptor. Comput Struct Biotechnol J. 2022;20:6503–11.
He X, Zhao L, Tian Y, Li R, Chu Q, Gu Z, et al. Highly accurate carbohydrate-binding site prediction with DeepGlycanSite. Nat Commun. 2024;15:5163.
Yim J, Stärk H, Corso G, Jing B, Barzilay R, Jaakkola TS. Diffusion models in protein structure and docking. Wiley Interdiscip Rev Comput Mol Sci. 2024;14:e1711.
Acknowledgments
This work was supported by National Natural Science Foundation of China (32130022 and 82121005 to HEX); the National Key R&D Program of China (2022YFC2703105 to HEX, 2019YFA0904200); CAS Strategic Priority Research Program (XDB37030103 to HEX); Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to HEX); the Lingang Laboratory (LG-GG-202204-01 to HEX); State Key Laboratory of Drug Research (SKLDR-2023-TT-04 to HEX).
Author information
Authors and Affiliations
Contributions
XHH, JRL, YSW and SNL deployed AlphaFold3 locally; SYS evaluated and analyzed the structures and data, and performed molecular docking; SYS, XHH and HEX wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shen, Sy., Li, Jr., Wang, Ys. et al. An update for AlphaFold3 versus experimental structures: assessing the precision of small molecule binding in GPCRs. Acta Pharmacol Sin (2025). https://doi.org/10.1038/s41401-025-01617-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41401-025-01617-4