Abstract
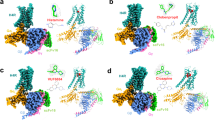
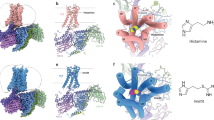
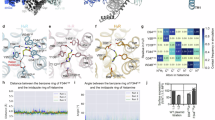
Histamine H3 receptor (H3R) and H4 receptor (H4R) are key members of the histamine receptor family, with H3R as a potential target for narcolepsy treatments and H4R as a candidate for next-generation antihistamines for inflammatory and allergic diseases. Although progress has been made in understanding the structure of histamine receptors, the detailed mechanisms of ligand recognition and receptor antagonism for H3R and H4R remain unclear. In this study, using cryo-electron microscopy, we present an inactive structure of H4R bound to a selective antagonist, adriforant, and two Gi-coupled structures of H3R and H4R in complex with histamine. Our structural and mutagenesis analyses provide insights into the selective binding of adriforant to H4R and the recognition of histamine across histamine receptors. Our findings also uncovered distinct antagonistic mechanisms for H3R and H4R and identified the role of aromatic amino acids on extracellular loop 2 in modulating the constitutive activity of H3R and H4R. These findings advance our knowledge of the functional modulation of histamine receptors, providing a foundation for the development of targeted therapeutics for neurological and immune-related disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Church MK. Allergy, histamine and antihistamines. Handb Exp Pharmacol. 2017;241:321–31.
Obrink KJ. Histamine and gastric acid secretion. A review. Scand J Gastroenterol Suppl. 1991;180:4–8.
Nuutinen S, Panula P. Histamine in neurotransmission and brain diseases. Adv Exp Med Biol. 2010;709:95–107.
Jutel M, Blaser K, Akdis CA. Histamine in allergic inflammation and immune modulation. Int Arch Allergy Immunol. 2005;137:82–92.
Booth RG, Moniri NH, Bakker RA, Choksi NY, Nix WB, Timmerman H, et al. A novel phenylaminotetralin radioligand reveals a subpopulation of histamine H1 receptors. J Pharmacol Exp Ther. 2002;302:328–36.
Beukers MW, Klaassen CH, De Grip WJ, Verzijl D, Timmerman H, Leurs R. Heterologous expression of rat epitope-tagged histamine H2 receptors in insect Sf9 cells. Br J Pharmacol. 1997;122:867–74.
Cogé F, Guénin SP, Audinot V, Renouard-Try A, Beauverger P, Macia C, et al. Genomic organization and characterization of splice variants of the human histamine H3 receptor. Biochem J. 2001;355:279–88.
Gbahou F, Vincent L, Humbert-Claude M, Tardivel-Lacombe J, Chabret C, Arrang JM. Compared pharmacology of human histamine H3 and H4 receptors: structure-activity relationships of histamine derivatives. Br J Pharmacol. 2006;147:744–54.
Lin JS, Sergeeva OA, Haas HL. Histamine H3 receptors and sleep-wake regulation. J Pharmacol Exp Ther. 2011;336:17–23.
Stevens DR, Eriksson KS, Brown RE, Haas HL. The mechanism of spontaneous firing in histamine neurons. Behav Brain Res. 2001;124:105–12.
Flik G, Folgering JH, Cremers TI, Westerink BH, Dremencov E. Interaction between brain histamine and serotonin, norepinephrine, and dopamine systems: in vivo microdialysis and electrophysiology study. J Mol Neurosci. 2015;56:320–8.
Schlicker E, Werthwein S, Zentner J. Histamine H3 receptor-mediated inhibition of noradrenaline release in the human brain. Fundam Clin Pharmacol. 1999;13:120–2.
Yamada M, Tokimasa T, Koketsu K. Effects of histamine on acetylcholine release in bullfrog sympathetic ganglia. Eur J Pharmacol. 1982;82:15–20.
Passani MB, Blandina P, Torrealba F. The histamine H3 receptor and eating behavior. J Pharmacol Exp Ther. 2011;336:24–9.
Passani MB, Lin JS, Hancock A, Crochet S, Blandina P. The histamine H3 receptor as a novel therapeutic target for cognitive and sleep disorders. Trends Pharmacol Sci. 2004;25:618–25.
Mommert S, Kleiner S, Gehring M, Eiz-Vesper B, Stark H, Gutzmer R, et al. Human basophil chemotaxis and activation are regulated via the histamine H4 receptor. Allergy. 2016;71:1264–73.
del Rio R, Noubade R, Saligrama N, Wall EH, Krementsov DN, Poynter ME, et al. Histamine H4 receptor optimizes T regulatory cell frequency and facilitates anti-inflammatory responses within the central nervous system. J Immunol. 2012;188:541–7.
Ehling S, Roßbach K, Dunston SM, Stark H, Bäumer W. Allergic inflammation is augmented via histamine H4 receptor activation: The role of natural killer cells in vitro and in vivo. J Dermatol Sci. 2016;83:106–15.
Schirmer B, Neumann D. The function of the histamine H4 receptor in inflammatory and inflammation-associated diseases of the gut. Int J Mol Sci. 2021;22.
Mehta P, Miszta P, Rzodkiewicz P, Michalak O, Krzeczyński P, Filipek S. Enigmatic histamine receptor H4 for potential treatment of multiple inflammatory, autoimmune, and related diseases. Life. 2020;10:50.
Marson CM. Targeting the histamine H4 receptor. Chem Rev. 2011;111:7121–56.
Arrang JM, Morisset S, Gbahou F. Constitutive activity of the histamine H3 receptor. Trends Pharmacol Sci. 2007;28:350–7.
Wifling D, Löffel K, Nordemann U, Strasser A, Bernhardt G, Dove S, et al. Molecular determinants for the high constitutive activity of the human histamine H4 receptor: functional studies on orthologues and mutants. Br J Pharmacol. 2015;172:785–98.
Keam SJ. Pitolisant: pediatric first approval. Paediatr Drugs. 2023;25:483–8.
Neumann D. Role of the histamine H4-receptor in bronchial asthma. Handb Exp Pharmacol. 2017;241:347–59.
Boyle DL, DePrimo SE, Calderon C, Chen D, Dunford PJ, Barchuk W, et al. Toreforant, an orally active histamine H(4)-receptor antagonist, in patients with active rheumatoid arthritis despite methotrexate: mechanism of action results from a phase 2, multicenter, randomized, double-blind, placebo-controlled synovial biopsy study. Inflamm Res. 2019;68:261–74.
Aldossari AA, Assiri MA, Ansari MA, Nadeem A, Attia SM, Bakheet SA, et al. Histamine H4 receptor antagonist ameliorates the progression of experimental autoimmune encephalomyelitis via regulation of T-cell imbalance. Int J Mol Sci. 2023;24:15273.
Tsuji G, Yamamura K, Kawamura K, Kido-Nakahara M, Ito T, Nakahara T. Novel therapeutic targets for the treatment of atopic dermatitis. Biomedicines. 2023;11:1303.
Werfel T, Layton G, Yeadon M, Whitlock L, Osterloh I, Jimenez P, et al. Efficacy and safety of the histamine H(4) receptor antagonist ZPL-3893787 in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143:1830–7.e4.
Kollmeier A, Francke K, Chen B, Dunford PJ, Greenspan AJ, Xia Y, et al. The histamine H4 receptor antagonist, JNJ 39758979, is effective in reducing histamine-induced pruritus in a randomized clinical study in healthy subjects. J Pharmacol Exp Ther. 2014;350:181–7.
Katoh N. Emerging treatments for atopic dermatitis. J Dermatol. 2021;48:152–7.
Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, Katritch V, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–76.
Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem Biol. 2016;11:400–8.
Duan J, Shen DD, Zhou XE, Bi P, Liu QF, Tan YX, et al. Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy. Nat Commun. 2020;11:4121.
Liu P, Jia MZ, Zhou XE, De Waal PW, Dickson BM, Liu B, et al. The structural basis of the dominant negative phenotype of the Gαi1β1γ2 G203A/A326S heterotrimer. Acta Pharmacol Sin. 2016;37:1259–72.
Tsutsumi N, Mukherjee S, Waghray D, Janda CY, Jude KM, Miao Y, et al. Structure of human Frizzled5 by fiducial-assisted cryo-EM supports a heterodimeric mechanism of canonical Wnt signaling. Elife. 2020;9:e58464.
Zhang K, Wu H, Hoppe N, Manglik A, Cheng Y. Fusion protein strategies for cryo-EM study of G protein-coupled receptors. Nat Commun. 2022;13:4366.
Peng X, Yang L, Liu Z, Lou S, Mei S, Li M, et al. Structural basis for recognition of antihistamine drug by human histamine receptor. Nat Commun. 2022;13:6105.
Kimura KT, Asada H, Inoue A, Kadji FMN, Im D, Mori C, et al. Structures of the 5-HT(2A) receptor in complex with the antipsychotics risperidone and zotepine. Nat Struct Mol Biol. 2019;26:121–8.
Shao Z, Yan W, Chapman K, Ramesh K, Ferrell AJ, Yin J, et al. Structure of an allosteric modulator bound to the CB1 cannabinoid receptor. Nat Chem Biol. 2019;15:1199–205.
Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–2.
Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–6.
Sanchez-Garcia R, Gomez-Blanco J, Cuervo A, Carazo JM, Sorzano COS, Vargas J. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun Biol. 2021;4:874.
Wu C, Xu Y, He Q, Li D, Duan J, Li C, et al. Ligand-induced activation and G protein coupling of prostaglandin F(2α) receptor. Nat Commun. 2023;14:2668.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12.
Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32.
Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat. 2004;11:53–5.
Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82.
Olsen RHJ, DiBerto JF, English JG, Glaudin AM, Krumm BE, Slocum ST, et al. TRUPATH, an open-source biosensor platform for interrogating the GPCR transducerome. Nat Chem Biol. 2020;16:841–9.
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–18.
Olsson MH, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput. 2011;7:525–37.
Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–65.
Panula P, Chazot PL, Cowart M, Gutzmer R, Leurs R, Liu WL, et al. International union of basic and clinical pharmacology. XCVIII. Histamine Receptors. Pharmacol Rev. 2015;67:601–55.
Shen Q, Tang X, Wen X, Cheng S, Xiao P, Zang SK, et al. Molecular determinant underlying selective coupling of primary G-protein by class A GPCRs. Adv Sci (Weinh). 2024;11:e2310120.
Nicoli A, Dunkel A, Giorgino T, de Graaf C, Di Pizio A. Classification model for the second extracellular loop of class A GPCRs. J Chem Inf Model. 2022;62:511–22.
Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27.
Mear Y, Enjalbert A, Thirion S. GHS-R1a constitutive activity and its physiological relevance. Front Neurosci. 2013;7:87.
Nijmeijer S, Leurs R, Vischer HF. Constitutive activity of the histamine H1 receptor. Methods Enzymol. 2010;484:127–47.
Wang D, Guo Q, Wu Z, Li M, He B, Du Y, et al. Molecular mechanism of antihistamines recognition and regulation of the histamine H1 receptor. Nat Commun. 2024;15:84.
Robertson MJ, Papasergi-Scott MM, He F, Seven AB, Meyerowitz JG, Panova O, et al. Structure determination of inactive-state GPCRs with a universal nanobody. Nat Struct Mol Biol. 2022;29:1188–95.
Xia R, Wang N, Xu Z, Lu Y, Song J, Zhang A, et al. Cryo-EM structure of the human histamine H(1) receptor/G(q) complex. Nat Commun. 2021;12:2086.
Köck Z, Schnelle K, Persechino M, Umbach S, Schihada H, Januliene D, et al. Cryo-EM structure of cell-free synthesized human histamine 2 receptor/G(s) complex in nanodisc environment. Nat Commun. 2024;15:1831.
Acknowledgements
The cryo-EM data were collected at the Advanced Center for Electron Microscopy, Shanghai Institute of Materia Medica. We thank all staff at the institution for their assistance with cryo-EM data collection. This work was partially supported by the National Key R&D Program of China (2022YFC2703105 to HEX); the Lingang Laboratory (LG-GG-202204-01 to YJ); the National Natural Science Foundation of China (32171187 to YJ; 82121005 to YJ, HEX and DHY; 32130022 to HEX; 82273985 to DHY; 82273961 to MWW); the CAS Strategic Priority Research Program (XDB37030103 to HEX); the Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to HEX); the National Key Basic Research Program of China (2023YFA1800804 to DHY); the Shanghai Municipality Science and Technology Development Fund (21JC1401600 to DHY); the Program of Shanghai Academic/Technology Research Leader (23XD1400900 to DHY); Sanya Science and Technology Innovation Special Project (2022KJCX77 to DHY).
Author information
Authors and Affiliations
Contributions
SSJ screened constructs and purification conditions, prepared the protein samples for final structure determination, participated in cryo-EM grid inspection and data collection, analyzed structures and performed some of the functional studies, and prepared the Figs. and wrote the initial manuscript. HZ performed cryo-EM data collection and structure determination, built and refined structure models, prepared Figs. and manuscript draft. JHY and XQC prepared the constructs for functional assays and conducted functional studies and functional data analysis. CRW. participated in protein sample optimization and purification, participated in grid preparation, cryo-EM grid inspection, and gave advice to the functional study. KW participated in data collection and analysis. DHY. and MWW conceived and supervised the functional study. HEX conceived and supervised the structure determination. YJ initiated, conceived, and supervised the project, analyzed structures, prepared the Figs. and wrote the manuscript with inputs from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jin, Ss., Zhang, H., Yan, Jh. et al. Decoding ligand recognition and constitutive activation of histamine H3 and H4 receptors. Acta Pharmacol Sin 47, 186–196 (2026). https://doi.org/10.1038/s41401-025-01633-4
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41401-025-01633-4