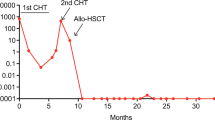
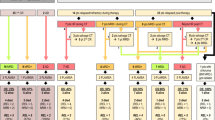
Abstract
NPM1 mutated acute myeloid leukemia (AML) comprises roughly 30% of all AML cases and is mainly classified as favorable or intermediate-risk according to the European Leukemia Net stratification. Some patients, however, either have a poor response to initial intensive chemotherapy or ultimately relapse. NPM1 mutations are common, generally stable at early relapse and AML specific, features which make them ideal targets for measurable residual disease (MRD) monitoring. MRD monitoring via molecular analysis during the course of treatment can inform the role of allogeneic stem cell transplantation (HCT) in first remission in patients with NPM1 mutated AML with high-risk co-occurring mutations, particularly FLT3-ITD, and in favorable risk patients who do not achieve defined molecular milestones. In this review, we evaluate the prognostic role of MRD monitoring in NPM1 mutated AML and its use as a predictive biomarker to refine risk stratification and inform decision making regarding treatment. We explore the impact of pre-HCT MRD positivity on post-HCT outcomes in this AML subset, and how HCT-related factors such as conditioning intensity may influence this risk.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21.
Sasaki K, Ravandi F, Kadia TM, DiNardo CD, Short NJ, Borthakur G, et al. De novo acute myeloid leukemia: a population-based study of outcome in the United States based on the surveillance, epidemiology, and end results (SEER) database, 1980 to 2017. Cancer. 2021;127:2049–61.
Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–33.
Falini B, Martelli MP, Bolli N, Sportoletti P, Liso A, Tiacci E, et al. Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood. 2011;117:1109–20.
Schlenk Richard F, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. New Eng J Med. 2008;358:1909–18.
Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–77.
Becker H, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Margeson D, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a cancer and leukemia group B study. J Clin Oncol. 2010;28:596–604.
Metzeler KH, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Görlich D, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–98.
Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–19.
Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. International consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140:1200–28.
Balsat M, Renneville A, Thomas X, de Botton S, Caillot D, Marceau A, et al. Postinduction minimal residual disease predicts outcome and benefit from allogeneic stem cell transplantation in acute myeloid leukemia with NPM1 mutation: a study by the acute leukemia French association group. J Clin Oncol. 2017;35:185–93.
Poiré X, Labopin M, Polge E, Blaise D, Chevallier P, Maertens J, et al. Hematopoietic stem cell transplantation for adult patients with isolated NPM1 mutated acute myeloid leukemia in first remission. Am J Hematol. 2019;94:231–9.
Borrow J, Dyer SA, Akiki S, Griffiths MJ. Molecular roulette: nucleophosmin mutations in AML are orchestrated through N-nucleotide addition by TdT. Blood. 2019;134:2291–303.
Falini B, Bolli N, Shan J, Martelli MP, Liso A, Pucciarini A, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107:4514–23.
Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–66.
Vassiliou GS, Cooper JL, Rad R, Li J, Rice S, Uren A, et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet. 2011;43:470–5.
Bolli N, Nicoletti I, De Marco MF, Bigerna B, Pucciarini A, Mannucci R, et al. Born to be exported: COOH-terminal nuclear export signals of different strength ensure cytoplasmic accumulation of nucleophosmin leukemic mutants. Cancer Res. 2007;67:6230–7.
Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA. 2014;111:2548–53.
Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559:400–4.
Loberg MA, Bell RK, Goodwin LO, Eudy E, Miles LA, SanMiguel JM, et al. Sequentially inducible mouse models reveal that NPM1 mutation causes malignant transformation of dnmt3a-mutant clonal hematopoiesis. Leukemia. 2019;33:1635–49.
Herudkova Z, Culen M, Folta A, Jeziskova I, Cerna J, Loja T, et al. Clonal hierarchy of main molecular lesions in acute myeloid leukaemia. Br J Haematol. 2020;190:562–72.
Potter N, Miraki-Moud F, Ermini L, Titley I, Vijayaraghavan G, Papaemmanuil E, et al. Single cell analysis of clonal architecture in acute myeloid leukaemia. Leukemia. 2019;33:1113–23.
Hubmann M, Köhnke T, Hoster E, Schneider S, Dufour A, Zellmeier E, et al. Molecular response assessment by quantitative real-time polymerase chain reaction after induction therapy in NPM1-mutated patients identifies those at high risk of relapse. Haematologica. 2014;99:1317–25.
Krönke J, Bullinger L, Teleanu V, Tschürtz F, Gaidzik VI, Kühn MW, et al. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood. 2013;122:100–8.
Garciaz S, Hospital MA, Saillard C, Hicheri Y, D’Incan E, Rey J, et al. IDH mutations identify a subgroup of NPM1 patients with a more favorable prognosis. a retrospective multicenter study of the filo group. Blood. 2020;136:39–40.
Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Krönke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–43.
Ley Timothy J, Ding L, Walter Matthew J, McLellan Michael D, Lamprecht T, Larson David E, et al. DNMT3A mutations in acute myeloid leukemia. New Eng J Med. 2010;363:2424–33.
Chou W-C, Chou S-C, Liu C-Y, Chen C-Y, Hou H-A, Kuo Y-Y, et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118:3803–10.
Döhner K, Schlenk RF, Habdank M, Scholl C, Rücker FG, Corbacioglu A, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106:3740–6.
Boddu P, Kantarjian H, Borthakur G, Kadia T, Daver N, Pierce S, et al. Co-occurrence of FLT3-TKD and NPM1 mutations defines a highly favorable prognostic AML group. Blood Adv. 2017;1:1546–50.
Liu J, Qin W, Wang B, Wang Z, Hua H, Zhou F, et al. PTPN11 mutations in adult acute myeloid leukaemia: Prevalence and clinical implications in the context of NPM1 mutation. Leuk Res. 2022;118:106859.
El Hussein S, DiNardo CD, Takahashi K, Khoury JD, Fang H, Furudate K, et al. Acquired WT1 mutations contribute to relapse of NPM1-mutated acute myeloid leukemia following allogeneic hematopoietic stem cell transplant. Bone Marrow Transplant. 2022;57:370–6.
Kayser S, Benner A, Thiede C, Martens U, Huber J, Stadtherr P, et al. Pretransplant NPM1 MRD levels predict outcome after allogeneic hematopoietic stem cell transplantation in patients with acute myeloid leukemia. Blood Cancer J. 2016;6:e449.
Ivey A, Hills RK, Simpson MA, Jovanovic JV, Gilkes A, Grech A, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374:422–33.
Heuser M, Freeman SD, Ossenkoppele GJ, Buccisano F, Hourigan CS, Ngai LL, et al. 2021 update on MRD in acute myeloid leukemia: a consensus document from the European LeukemiaNet MRD Working Party. Blood. 2021;138:2753–67.
Short NJ, Fu C, Berry DA, Walter RB, Freeman SD, Hourigan CS, et al. Association of hematologic response and assay sensitivity on the prognostic impact of measurable residual disease in acute myeloid leukemia: a systematic review and meta-analysis. Leukemia. 2022;36:2817–26.
Hourigan CS, Dillon LW, Gui G, Logan BR, Fei M, Ghannam J, et al. Impact of conditioning intensity of allogeneic transplantation for acute myeloid leukemia with genomic evidence of residual disease. J Clin Oncol. 2020;38:1273–83.
Pratz KW, Jonas BA, Pullarkat V, Recher C, Schuh AC, Thirman MJ, et al. Measurable residual disease response and prognosis in treatment-naïve acute myeloid leukemia with venetoclax and azacitidine. J Clin Oncol. 2021;40:855–65.
Krönke J, Schlenk RF, Jensen KO, Tschürtz F, Corbacioglu A, Gaidzik VI, et al. Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: a study from the German-Austrian acute myeloid leukemia study group. J Clin Oncol. 2011;29:2709–16.
Kapp-Schwoerer S, Weber D, Corbacioglu A, Gaidzik VI, Paschka P, Krönke J, et al. Impact of gemtuzumab ozogamicin on MRD and relapse risk in patients with NPM1-mutated AML: results from the AMLSG 09-09 trial. Blood. 2020;136:3041–50.
Jongen-Lavrencic M, Grob T, Hanekamp D, Kavelaars François G, al Hinai A, Zeilemaker A, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med. 2018;378:1189–99.
Othman J, Tiong IS, O’Nions J, Dennis M, Mokretar K, Ivey A, et al. Molecular MRD is strongly prognostic in patients with NPM1-mutated AML receiving venetoclax-based nonintensive therapy. Blood. 2024;143:336–41.
Pettersson L, Johansson Alm S, Almstedt A, Chen Y, Orrsjö G, Shah-Barkhordar G, et al. Comparison of RNA- and DNA-based methods for measurable residual disease analysis in NPM1-mutated acute myeloid leukemia. Int J Lab Hematol. 2021;43:664–74.
Salipante SJ, Fromm JR, Shendure J, Wood BL, Wu D. Detection of minimal residual disease in NPM1-mutated acute myeloid leukemia by next-generation sequencing. Mod Pathol. 2014;27:1438–46.
Mencia-Trinchant N, Hu Y, Alas MA, Ali F, Wouters BJ, Lee S, et al. Minimal residual disease monitoring of acute myeloid leukemia by massively multiplex digital PCR in patients with NPM1 mutations. J Mol Diagn. 2017;19:537–48.
Stahl T, Böhme MU, Kröger N, Fehse B. Digital PCR to assess hematopoietic chimerism after allogeneic stem cell transplantation. Exp Hematol. 2015;43:462–8. e1
Huggett JF, Whale A. Digital PCR as a novel technology and its potential implications for molecular diagnostics. Clin Chem. 2013;59:1691–3.
Höllein A, Meggendorfer M, Dicker F, Jeromin S, Nadarajah N, Kern W, et al. NPM1 mutated AML can relapse with wild-type NPM1: persistent clonal hematopoiesis can drive relapse. Blood Adv. 2018;2:3118–25.
Cocciardi S, Dolnik A, Kapp-Schwoerer S, Rücker FG, Lux S, Blätte TJ, et al. Clonal evolution patterns in acute myeloid leukemia with NPM1 mutation. Nat Commun. 2019;10:2031.
Patkar N, Kodgule R, Kakirde C, Raval G, Bhanshe P, Joshi S, et al. Clinical impact of measurable residual disease monitoring by ultradeep next generation sequencing in NPM1 mutated acute myeloid leukemia. Oncotarget. 2018;9:36613–24.
Dillon LW, Gui G, Page KM, Ravindra N, Wong ZC, Andrew G, et al. DNA sequencing to detect residual disease in adults with acute myeloid leukemia prior to hematopoietic cell transplant. JAMA. 2023;329:745–55.
Zhou Y, Othus M, Walter RB, Estey EH, Wu D, Wood BL. Deep NPM1 sequencing following allogeneic hematopoietic cell transplantation improves risk assessment in adults with NPM1-mutated AML. Biol Blood Marrow Transpl. 2018;24:1615–20.
Schuurhuis GJ, Heuser M, Freeman S, Béné MC, Buccisano F, Cloos J, et al. Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD working party. Blood. 2018;131:1275–91.
Delsing Malmberg E, Johansson Alm S, Nicklasson M, Lazarevic V, Ståhlman S, Samuelsson T, et al. Minimal residual disease assessed with deep sequencing of NPM1 mutations predicts relapse after allogeneic stem cell transplant in AML. Leuk Lymphoma. 2019;60:409–17.
Schnittger S, Kern W, Tschulik C, Weiss T, Dicker F, Falini B, et al. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood. 2009;114:2220–31.
Lambert J, Lambert J, Nibourel O, Pautas C, Hayette S, Cayuela JM, et al. MRD assessed by WT1 and NPM1 transcript levels identifies distinct outcomes in AML patients and is influenced by gemtuzumab ozogamicin. Oncotarget. 2014;5:6280–8.
Shayegi N, Kramer M, Bornhäuser M, Schaich M, Schetelig J, Platzbecker U, et al. The level of residual disease based on mutant NPM1 is an independent prognostic factor for relapse and survival in AML. Blood. 2013;122:83–92.
Tiong IS, Dillon R, Ivey A, Kuzich JA, Thiagarajah N, Sharplin KM, et al. Clinical impact of NPM1-mutant molecular persistence after chemotherapy for acute myeloid leukemia. Blood Adv. 2021;5:5107–11.
Scott S, Dillon R, Thiede C, Sadiq S, Cartwright A, Clouston HJ, et al. Assessment of acute myeloid leukemia molecular measurable residual disease testing in an interlaboratory study. Blood Adv. 2023;7:3686–94.
Angenendt L, Röllig C, Montesinos P, Martínez-Cuadrón D, Barragan E, García R, et al. Chromosomal abnormalities and prognosis in NPM1-mutated acute myeloid leukemia: a pooled analysis of individual patient data from nine international cohorts. J Clin Oncol. 2019;37:2632–42.
Eckardt J-N, Bill M, Rausch C, Metzeler K, Spiekermann K, Stasik S, et al. Secondary-type mutations do not impact outcome in NPM1-mutated acute myeloid leukemia – implications for the European LeukemiaNet risk classification. Leukemia. 2023;37:2282–5.
Wright MF, Pozdnyakova O, Hasserjian RP, Aggarwal N, Shaver AC, Weinberg OK, et al. Secondary-type mutations do not impact prognosis in acute myelogenous leukemia AML with mutated NPM1. Am J Hematol. 2022;97:E462–5.
Wang Y, Quesada AE, Zuo Z, Medeiros LJ, Yin CC, Li S, et al. The impact of mutation of myelodysplasia-related genes in de novo acute myeloid leukemia carrying NPM1 mutation. Cancers. 2022;15:198.
Zhao D, Zarif M, Eladl E, Capo-Chichi JM, Smith AC, Atenafu EG, et al. NPM1-mutated AML-MRC diagnosed on the basis of history of MDS or MDS/MPN frequently harbours secondary-type mutations and confers inferior outcome compared to AML with mutated NPM1. Leuk Res. 2022;118:106869.
Othman J, Potter N, Ivey A, Jovanovic J, Freeman SD, Gilkes A, et al. The benefit of allogeneic transplant in 1st complete remission in NPM1 mutated AML with or without FLT3 ITD is restricted to those testing MRD positive after induction - an analysis of the UK NCRI AML17 and AML19 studies. Blood. 2023;142:425.
Lussana F, Caprioli C, Stefanoni P, Pavoni C, Spinelli O, Buklijas K, et al. Molecular detection of minimal residual disease before allogeneic stem cell transplantation predicts a high incidence of early relapse in adult patients with NPM1 positive acute myeloid leukemia. Cancers. 2019;11:1455.
Hourigan CS, Dillon LW, Gui G, Ravindra N, Wong Z, Andrew G, et al. Pre-MEASURE: multicenter evaluation of the prognostic significance of measurable residual disease testing prior to allogeneic transplantation for adult patients with AML in first remission. J Clin Oncol. 2022;40:7006.
Dillon R, Hills R, Freeman S, Potter N, Jovanovic J, Ivey A, et al. Molecular MRD status and outcome after transplantation in NPM1-mutated AML. Blood. 2020;135:680–8.
Fraccaroli A, Stauffer E, Koch K, Hirschbühl K, Breitkopf S, Ziemann F, et al. Presence of measurable residual disease in NPM1 positive AML in remission pre-transplant does not impact on survival after allogeneic hematopoietic stem cell transplantation. Blood. 2022;140:4882–3.
Jimenez-Chillon C, Othman J, Taussig D, Jimenez-Vicente C, Martinez-Roca A, Tiong IS, et al. Venetoclax–based low intensity therapy in molecular failure of NPM1-mutated AML. Blood Adv. 2024;8:343–52.
Tiong IS, Dillon R, Ivey A, Teh TC, Nguyen P, Cummings N, et al. Venetoclax induces rapid elimination of NPM1 mutant measurable residual disease in combination with low-intensity chemotherapy in acute myeloid leukaemia. Br J Haematol. 2021;192:1026–30.
Tiong IS, Hiwase DK, Abro E, Bajel A, Palfreyman E, Beligaswatte A, et al. Targeting molecular measurable residual disease and low-blast relapse in AML with venetoclax and low-dose cytarabine: a prospective phase II study (VALDAC). J Clin Oncol. 2024;42:2161–73.
Bastos-Oreiro M, Perez-Corral A, Martínez-Laperche C, Bento L, Pascual C, Kwon M, et al. Prognostic impact of minimal residual disease analysis by flow cytometry in patients with acute myeloid leukemia before and after allogeneic hemopoietic stem cell transplantation. Eur J Haematol. 2014;93:239–46.
Bacher U, Badbaran A, Fehse B, Zabelina T, Zander AR, Kröger N. Quantitative monitoring of NPM1 mutations provides a valid minimal residual disease parameter following allogeneic stem cell transplantation. Exp Hematol. 2009;37:135–42.
Bill M, Grimm J, Jentzsch M, Kloss L, Goldmann K, Schulz J, et al. Digital droplet PCR-based absolute quantification of pre-transplant NPM1 mutation burden predicts relapse in acute myeloid leukemia patients. Ann Hematol. 2018;97:1757–65.
Author information
Authors and Affiliations
Contributions
Contribution: MTN and MRC. performed literature review and wrote the manuscript. JLR edited the manuscript and contributed to the figures.
Corresponding author
Ethics declarations
Competing interests
MRC and MTN have no disclosures. JLR reports serving as a consultant and/or advisory board member for Sanofi and Bristol Myers Squibb; research support from Pfizer.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Christopher, M.R., Nawas, M.T. & Reagan, J.L. Molecular measurable residual disease monitoring and transplant indications in NPM1 mutated acute myeloid leukemia. Bone Marrow Transplant 60, 135–143 (2025). https://doi.org/10.1038/s41409-024-02465-2
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41409-024-02465-2