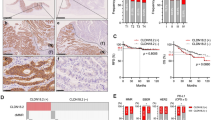
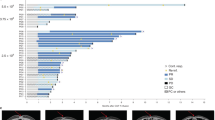
Abstract
Background
Claudin 18 isoform 2 (CLDN18.2) is a potential therapeutic target in gastric cancer (GC). However, combining chemotherapy with anti-CLDN18.2 antibodies has shown limited efficacy in CLDN18.2-positive GC, and chemotherapy-induced changes in the tumor microenvironment (TME) remain unclear.
Methods
This study analyzed 37 GC samples, including 11 CLDN18.2-positive cases, using single-cell RNA sequencing and multiplex immunofluorescence to assess chemotherapy-driven TME changes in CLDN18.2-positive GC.
Results
In chemotherapy-treated CLDN18.2-positive GC, cytotoxic natural killer (NK) cells displayed antibody-dependent cytotoxicity (ADCC)-related genes at lower levels than in untreated CLDN18.2-positive GC, while regulatory T cells (Tregs) and tumor-associated macrophages (TAMs) showed TGFB1 expression at higher levels. Additionally, NK cells, Tregs, and TAMs were more abundant in chemotherapy-treated than untreated CLDN18.2-positive GC. These chemotherapy-induced changes were absent in CLDN18.2-negative GC. Cell-cell interaction analysis identified unique interactions in chemotherapy-treated CLDN18.2-positive GC, including CCL5-CCR5 signaling between cytotoxic NK cells (Sender) and effector Tregs (Receptor) and TGFB1-TGFBR signaling between effector Tregs (Sender) and TAMs (Receptor). Cytotoxic NK cells expressed CCL5 at higher levels, CCR5-positive Tregs were more prevalent, and TAMs exhibited higher TGF-β receptor signature scores in chemotherapy-treated than untreated CLDN18.2-positive GC.
Conclusions
Our findings indicate that chemotherapy can drive immunosuppressive TME modifications specific to CLDN18.2-positive GC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The scRNA-seq data used in this study are available in the GEO database under accession code GSE268238. One case was excluded due to the absence of residual cancer cells in the resected specimen. Additional data, including code information, are available within the article, its supplementary information, or upon request from the authors.
Code availability
The scRNA-seq data used in this study are available in the GEO database under accession code GSE268238. One case was excluded due to the absence of residual cancer cells in the resected specimen. Additional data, including code information, are available within the article, its supplementary information, or upon request from the authors.
References
Shitara K, Özgüroğlu M, Bang YJ, Di Bartolomeo M, Mandalà M, Ryu MH, et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet. 2018;392:123–33.
Boku N, Yamamoto S, Fukuda H, Shirao K, Doi T, Sawaki A, et al. Fluorouracil versus combination of irinotecan plus cisplatin versus S-1 in metastatic gastric cancer: a randomised phase 3 study. Lancet Oncol. 2009;10:1063–9.
Koizumi W, Narahara H, Hara T, Takagane A, Akiya T, Takagi M, et al. S-1 plus cisplatin versus S-1 alone for fi rst-line treatment of advanced gastric cancer (SPIRITS trial): a phase III trial. Lancet Oncol. 2008;9:215–21.
Kang YK, Chen LT, Ryu MH, Oh DY, Oh SC, Chung HC, et al. Nivolumab plus chemotherapy versus placebo plus chemotherapy in patients with HER2-negative, untreated, unresectable advanced or recurrent gastric or gastro-oesophageal junction cancer (ATTRACTION-4): a randomised, multicentre, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2022;23:234–47.
Marabelle A, Le DT, Ascierto PA, Giacomo Anna, Di M, De Jesus-Acosta A, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/ mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. 2019;38:1.
Bang KoreaS, Bang Y-J, Cutsem Y-J, Van E, Feyereislova A, Chung HC, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97.
National Cancer Institute Surveillance Epidemiology and End Results Program. Cancer Stat Facts: Stomach Cancer.https://seer.cancer.gov/statfacts/html/stomach.html.Accessed September 2024.
Sahin U, Koslowski M, Dhaene K, Usener D, Brandenburg G, Seitz G, et al. Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin Cancer Res. 2008;14:7624–34.
Shah MA, Shitara K, Ajani JA, Bang YJ, Enzinger P, Ilson D, et al. Zolbetuximab plus CAPOX in CLDN18.2-positive gastric or gastroesophageal junction adenocarcinoma: the randomized, phase 3 GLOW trial. Nat Med. 2023;29:2133–41.
Kubota Y, Kawazoe A, Mishima S, Nakamura Y, Kotani D, Kuboki Y, et al. Comprehensive clinical and molecular characterization of claudin 18.2 expression in advanced gastric or gastroesophageal junction cancer. ESMO Open. 2023;8:e100762.
Shitara K, Lordick F, Bang YJ, Enzinger P, Ilson D, Shah MA, et al. Zolbetuximab plus mFOLFOX6 in patients with CLDN18.2-positive, HER2-negative, untreated, locally advanced unresectable or metastatic gastric or gastro-oesophageal junction adenocarcinoma (SPOTLIGHT): a multicentre, randomised, double-blind, phase 3 trial. Lancet. 2023;401:1655–68.
Dottermusch M, Krüger S, Behrens HM, Halske C, Röcken C. Expression of the potential therapeutic target claudin-18.2 is frequently decreased in gastric cancer: results from a large Caucasian cohort study. Virchows Arch. 2019;475:563–71.
Liu S, Zhang Z, Jiang L, Zhang M, Zhang C, Shen L. Claudin-18.2 mediated interaction of gastric cancer cells and cancer-associated fibroblasts drives tumor progression. Cell Commun Signal. 2024;22:27.
Niimi T, Nagashima K, Ward JM, Minoo P, Zimonjic DB, Popescu NC, et al. Claudin-18, a novel downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor, encodes lung- and stomach-specific isoforms through alternative splicing. Mol Cell Biol. 2001;21:7380–90.
Jia K, Chen Y, Sun Y, Hu Y, Jiao L, Ma J, et al. Multiplex immunohistochemistry defines the tumor immune microenvironment and immunotherapeutic outcome in CLDN18.2-positive gastric cancer. BMC Med. 2022;20:e369.
Sahin U, Schuler M, Richly H, Bauer S, Krilova A, Dechow T, et al. A phase I dose-escalation study of IMAB362 (Zolbetuximab) in patients with advanced gastric and gastro-oesophageal junction cancer. Eur J Cancer. 2018;100:17–26.
Ӧzlem Türeci, Mitnacht-Kraus R, Wöll S, Yamada T, Sahin U. Characterization of zolbetuximab in pancreatic cancer models. Oncoimmunology. 2019;8:e1523096.
Luoma AM, Suo S, Williams HL, Sharova T, Sullivan K, Manos M, et al. Molecular pathways of colon inflammation induced by cancer immunotherapy. Cell. 2020;182:655–71.e22.
Yamada Y, Yamamoto T, Tsutsumi C, Matsumoto T, Noguchi S, Shimada Y, et al. Immature stroma and high infiltration of CD15+ cells are predictive markers of poor prognosis in different subsets of patients with pancreatic cancer. Cancer Sci. 2024;115:1001–13.
Okuda S, Ohuchida K, Nakamura S, Tsutsumi C, Hisano K, Mochida Y, et al. Neoadjuvant chemotherapy enhances anti-tumor immune response of tumor microenvironment in human esophageal squamous cell carcinoma. iScience. 2023;26:106480.
Tsutsumi C, Ohuchida K, Katayama N, Yamada Y, Nakamura S, Okuda S, et al. Tumor-infiltrating monocytic myeloid-derived suppressor cells contribute to the development of an immunosuppressive tumor microenvironment in gastric cancer. Gastric Cancer. 2024;27:248–62.
Nestorowa S, Hamey FK, Sala BP, Diamanti E, Shepherd M, Laurenti E, et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128:e20–31.
Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296.
Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–96.
McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–37.e4.
Attig S, Hennenlotter J, Pawelec G, Klein G, Koch SD, Pircher H, et al. Simultaneous infiltration of polyfunctional effector and suppressor T cells into renal cell carcinomas. Cancer Res. 2009;69:8412–9.
Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-β gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059–67.
Xiong D, Wang Y, You M. A gene expression signature of TREM2hi macrophages and γδ T cells predicts immunotherapy response. Nat Commun. 2020;11:5084.
Chen Y, Song Y, Du W, Gong L, Chang H, Zou Z. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci. 2019;26:78.
Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–60.
Singh P, Toom S, Huang Y. Anti-claudin 18.2 antibody as new targeted therapy for advanced gastric cancer. J Hematol Oncol. 2017;10:105.
Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, et al. Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med. 2006;203:1033–43.
Cózar B, Greppi M, Carpentier S, Narni-Mancinelli E, Chiossone L, Vivier E. Tumor-infiltrating natural killer cells. Cancer Discov. 2021;11:34–44.
Barao I, Hanash AM, Hallett W, Welniak LA, Sun K, Redelman D, et al. Suppression of natural killer cell-mediated bone marrow cell rejection by CD4 CD25 regulatory T cells. Proc Natl Acad Sci USA. 2006;103:5460–5.
Rossana T, Jessica DC, Jianhua Y, David C, Brittany T, Xiaoli Z. TGF-beta utilizes SMAD3 to inhibit CD16-mediated IFN-gamma production and antibody-dependent cellular cytotoxicity in human NK cells. J Immunol. 2008;181:3784–92.
Massagué J. TGFβ in cancer. Cell. 2008;134:215–30.
Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression — implications for anticancer therapy. Nat Rev Clin Oncol. 2019;16:356–71.
van Hooren L, Handgraaf SM, Kloosterman DJ, Karimi E, van Mil LWHG, Gassama AA, et al. CD103+ regulatory T cells underlie resistance to radio-immunotherapy and impair CD8+ T cell activation in glioblastoma. Nat Cancer. 2023;4:665–81.
Pan Y, Yu Y, Wang X, Zhang T. Tumor-associated macrophages in tumor immunity. Front Immunol. 2020;11:583084.
Mould KJ, Jackson ND, Henson PM, Seibold M, Janssen WJ. Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight. 2019;4:1062–75.
Li H, Miao Y, Zhong L, Feng S, Xu Y, Tang L, et al. Identification of TREM2-positive tumor-associated macrophages in esophageal squamous cell carcinoma: implication for poor prognosis and immunotherapy modulation. Front Immunol. 2023;14:1162032.
Ni C, Yang L, Xu Q, Yuan H, Wang W, Xia W, et al. CD68- and CD163-positive tumor infiltrating macrophages in non-metastatic breast cancer: A retrospective study and meta-analysis. J Cancer. 2019;10:4463–72.
Kyrysyuk O, Wucherpfennig KW. Designing cancer immunotherapies that engage T cells and NK cells. Annu Rev Immunol. 2023;41:17–38.
De Sanctis F, Dusi S, Caligola S, Anselmi C, Petrova V, Rossi B, et al. Expression of the membrane tetraspanin claudin 18 on cancer cells promotes T lymphocyte infiltration and antitumor immunity in pancreatic cancer. Immunity. 2024;57:1378–93.
Luo J, Cheng K, Ji X, Gao C, Zhu R, Chen J, et al. Anlotinib enhanced CD8+ T cell infiltration via induction of CCL5 improves the efficacy of PD-1/PD-L1 blockade therapy in lung cancer. Cancer Lett. 2024;591:216892.
Seo W, Shimizu K, Kojo S, Okeke A, Kohwi-Shigematsu T, Fujii S, ichiro, et al. Runx-mediated regulation of CCL5 via antagonizing two enhancers influences immune cell function and anti-tumor immunity. Nat Commun. 2020;11:1562.
Yang X, Lin Y, Shi Y, Li B, Liu W, Yin W, et al. FAP Promotes immunosuppression by cancer-associated fibroblasts in the tumor microenvironment via STAT3-CCL2 Signaling. Cancer Res. 2016;76:4124–35.
Chen Z, Ding C, Chen J, Zheng S, Li Q. Pan-cancer analysis revealing the multidimensional expression and prognostic and immunologic roles of TGFB1 in cancer. J Int Med Res. 2024;52:3000605231221361.
Zhang F, Wang H, Wang X, Jiang G, Liu H, Zhang G, et al. TGF-β induces M2-like macrophage polarization via SNAIL-mediated suppression of a pro-inflammatory phenotype. Oncotarget. 2016;7:52294–306.
Oliveira G, Stromhaug K, Klaeger S, Kula T, Frederick DT, Le PM, et al. Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature. 2021;596:119–25.
Huang D, Liu X, Gao X, Choi CK, Giglio G, Farah L, et al. Meteorin-like protein/METRNL/Interleukin-41 ameliorates atopic dermatitis-like inflammation. Allergy. 2025;80:474–88.
Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–8.
Cao W, Xing H, Li Y, Tian W, Song Y, Jiang Z, et al. Claudin18.2 is a novel molecular biomarker for tumor-targeted immunotherapy. Biomark Res. 2022;10:38.
Woan KV, Miller JS. Harnessing natural killer cell antitumor immunity: from the bench to bedside. Cancer Immunol Res. 2019;7:1742–7.
Fabian KP, Wolfson B, Hodge JW. From immunogenic cell death to immunogenic modulation: select chemotherapy regimens induce a spectrum of immune-enhancing activities in the tumor microenvironment. Front Oncol. 2021;11:728018.
Tsutsumi C, Ohuchida K, Tsutsumi H, Shimada Y, Yamada Y, Son K, et al. TIM3 on natural killer cells regulates antibody-dependent cellular cytotoxicity in HER2-positive gastric cancer. Cancer Lett. 2025;611:217412.
Zeng Z, Lan T, Wei Y, Wei X. CCL5/CCR5 axis in human diseases and related treatments. Genes Dis. 2022;9:12–27.
Zaiatz-Bittencourt V, Finlay DK, Gardiner CM. Canonical TGF-β signaling pathway represses human NK cell metabolism. J Immunol. 2018;200:3934–41.
Bergsland CH, Jeanmougin M, Moosavi SH, Svindland A, Bruun J, Nesbakken A, et al. Spatial analysis and CD25-expression identify regulatory T cells as predictors of a poor prognosis in colorectal cancer. Mod Pathol. 2022;35:1236–46.
Acknowledgements
We thank E. Manabe and S. Sadatomi of Kyushu University for their expert technical assistance.
Funding
This study was funded by JSPS KAKENHI (Grant Number JP22H00480, JP23K27461, JP23K08175, JP24K11743, and JP24K11849) and JSPS Research Fellow PD (JP23KJ1698).
Author information
Authors and Affiliations
Contributions
CT designed the experimental approach, performed the experimental work, analyzed data, coordinated projects, and wrote the manuscript. KO designed the experimental approach, analyzed data, wrote the manuscript, coordinated projects, and contributed to data interpretation and discussion. CT, MI, KS, YM and NT performed the experimental work. MI, KS, YM, CI, NK, YM, NI, KN and HO contributed to data interpretation and discussion. KO, KH and KS provided and prepared human samples. Immunohistochemistry was analyzed by CT, YY, YS and YO. MN coordinated projects, wrote the manuscript, and contributed to data interpretation and discussion.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods in this study were performed in accordance with the relevant guidelines and regulations. Specimens were collected from patients who underwent gastrectomy at Kyushu University Hospital, with written informed consent obtained between 2019 and 2023. Ethics approval for this study was obtained from the Kyushu University Ethics Committee (approval numbers: 2023-79 and 22002-00).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tsutsumi, C., Ohuchida, K., Yamada, Y. et al. Claudin18.2-positive gastric cancer-specific changes in neoadjuvant chemotherapy-driven immunosuppressive tumor microenvironment. Br J Cancer 132, 793–804 (2025). https://doi.org/10.1038/s41416-025-02981-y
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41416-025-02981-y