Abstract
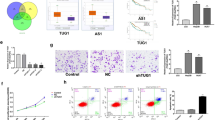
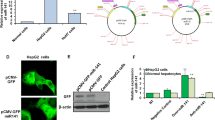
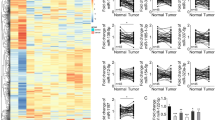
The aim of this study was to investigate the effect of lncRNA KCNQ1OT1 on HCC and to explore the possible underlying mechanisms. The expression levels of KCNQ1OT1, miR-149 and S1PR1 were detected by qRT-PCR assay. A dual luciferase reporter assay was used to detect the interaction between KCNQ1OT1 and miR-149, as well as miR-149 and S1PR1. The interaction between KCNQ1OT1 and miR-149 was further investigated by RNA pull-down assay. Wound healing assays and Transwell assays were carried out to determine cell migration and invasion. A xenograft tumour assay was used to validate the role of KCNQ1OT1 in vivo. KCNQ1OT1 and S1PR1 were significantly increased, but miR-149 was decreased in HCC cells. Luciferase reporter assays and RNA pull-down assays revealed that KCNQ1OT1 directly targeted miR-149. In addition, miR-149 bound to the 3’-UTR of S1PR1. Knockdown of KCNQ1OT1 or overexpression of miR-149 inhibited the invasion and migration of HCC cells. However, suppression of miR-149 could abrogate the effect of KCNQ1OT1 knockdown on the invasion and migration abilities of HCC cells. In vivo assays showed that KCNQ1OT1 knockdown suppressed tumour growth. This work suggests that lncRNA KCNQ1OT1 might act as a potential therapeutic target in HCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article.
References
Toro A, Ardiri A, Mannino M, Arcerito MC, Mannino G, Palermo F, et al. Effect of pre- and post-treatment α-fetoprotein levels and tumor size on survival of patients with hepatocellular carcinoma treated by resection, transarterial chemoembolization or radiofrequency ablation: a retrospective study. BMC Surg. 2014;14:40.
Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22:366–76.
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, et al. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–7.
Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61.
Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–400.
Zhu B, Cheng XB, Jiang YL, Cheng M, Chen LP, Bao JJ, et al. Silencing of KCNQ1OT1 decreases oxidative stress and pyroptosis of renal tubular epithelial cells. Diabetes Metab Syndr Obes. 2020;13:365–75.
Ren Y, Gao XP, Liang H, Zhang H, Hu CY. LncRNA KCNQ1OT1 contributes to oxygen-glucose-deprivation/reoxygenation-induced injury via sponging miR-9 in cultured neurons to regulate MMP8. Exp Mol Pathol. 2020;112:10435.
Zhang S, Ma H, Zhang D, Xie S, Wang W, Li Q, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9:742.
Li C, Miao R, Zhang J, Qu K, Liu C. Long non-coding RNA KCNQ1OT1 mediates the growth of hepatocellular carcinoma by functioning as a competing endogenous RNA of miR-504. Int J Oncol. 2018;52:1603–12.
Zhang Y, Guo X, Xiong L, Yu L, Li Z, Guo Q, et al. Comprehensive analysis of microRNA-regulated protein interaction network reveals the tumor suppressive role of microRNA-149 in human hepatocellular carcinoma via targeting Akt-mTOR pathway. Mol Cancer. 2014;13:253.
Liu XX, Wang M, Xu D, Yang JH, Kang HF, Wang XJ, et al. Quantitative assessment of the association between genetic variants in microRNAs and colorectal cancer risk. Biomed Res Int. 2015;2015:1–10.
Izquierdo L, Ingelmo-Torres M, Mallofrã C, Lozano JJ, Verhasselt-Crinquette M, Leroy X, et al. Prognostic value of microRNA expression pattern in upper tract urothelial carcinoma. BJU Int. 2014;113:813–21.
Xu Q, Liu JW, Yuan Y. Comprehensive assessment of the association between miRNA polymorphisms and gastric cancer risk. Mutat Res Rev Mutat Res. 2015;763:148–60.
Wang C, Mao J, Redfield S, Mo Y, Lage JM, Zhou X. Systemic distribution, subcellular localization and differential expression of sphingosine-1-phosphate receptors in benign and malignant human tissues. Exp Mol Pathol. 2014;97:259–65.
Zhou P, Huang G, Zhao Y, Zhong D, Xu Z, Zeng Y, et al. MicroRNA-363-mediated downregulation of S1PR1 suppresses the proliferation of hepatocellular carcinoma cells. Cell Signal. 2014;26:1347–54.
Zhang Y, Guo X, Xiong L, Kong X, Xu Y, Liu C, et al. MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett. 2012;586:4362–70.
Lin ZY, Huang YQ, Zhang YQ, Han ZD, He HC, Ling XH, et al. MicroRNA-224 inhibits progression of human prostate cancer by downregulating TRIB1. Int J Cancer. 2014;135:541–50.
Zhang Z, Zhang Y, Sun XX, Ma X, Chen ZN. MicroRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol Cancer. 2015;14:5.
Jiang R, Zhao C, Wang X, Wang S, Sun X, Tian Y, et al. Resistin-like molecule-β promotes invasion and migration of gastric carcinoma cells. Med Sci Monit. 2016;22:937–42.
Yuen MF, Hou JL, Chutaputti A. Hepatocellular carcinoma in the Asia Pacific region. J Gastroenterol Hepatol. 2010;24:346–53.
Chi HC, Tsai CY, Tsai MM, Yeh CT, Lin KH. Roles of long noncoding RNAs in recurrence and metastasis of radiotherapy-resistant cancer stem cells. Int J Mol Sci. 2017;18:1903.
Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603.
Bartel D. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Kwak PB, Iwasaki S, Tomari Y. The microRNA pathway and cancer. Cancer Sci. 2010;101:2309–15.
Kirschner MB, Cheng YY, Armstrong NJ, Lin RCY, Kao SC, Linton A, et al. MiR-score: a novel 6-microRNA signature that predicts survival outcomes in patients with malignant pleural mesothelioma. Mol Oncol. 2015;9:715–26.
Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, et al. Prognostic and predictive value of a microRNA signature in stage Ii colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–306.
Bagnoli M, Canevari S, Califano D, Losito S, Maio MD, Raspagliesi F, et al. Development and validation of a microRNA-based signature (MiROvaR) to predict early relapse or progression of epithelial ovarian cancer: a cohort study. Lancet Oncol. 2016;17:1137–46.
Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57.
Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–47.
Wong CC, Wong C, Tung EK, Au SL, Lee JM, Poon RT, et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140:322–31.
Yang H, Fang F, Chang R, Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor β receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205–17.
Qu J, Li M, Zhong W, Hu C. Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int J Clin Exp Med. 2015;8:17110.
Zhao N, Sun H, Sun B, Zhu D, Zhao X, Wang Y, et al. Mir-27a-3p suppresses tumor metastasis and VM by down-regulating VE-cadherin expression and inhibiting EMT: an essential role for Twist-1 in HCC. Sci Rep. 2016;6:23091.
Tania M, Khan MA, Fu J, Tania M, Khan MA, Fu J. Epithelial to mesenchymal transition inducing transcription factors and metastatic cancer. Tumor Biol. 2014;35:7335–42.
Battistelli C, Cicchini C, Santangelo L, Tramontano A, Grassi L, Gonzalez FJ, et al. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36:942–55.
Tang JW, Xie Y, Xu XL, Yin Y, Jiang RQ, Deng L, et al. Bidirectional transcription of Linc00441 and RB1 via H3K27 modification-dependent way promotes hepatocellular carcinoma. Cell Death Dis. 2017;8:e2675.
Ando H, Okamoto A, Yokota M, Shimizu K, Asai T, Dewa T, et al. Development of a miR-92a delivery system for anti-angiogenesis-based cancer therapy. J Gene Med. 2013;15:20–7.
Shigoka M, Tsuchida A, Matsudo T, Nagakawa Y, Saito H, Suzuki Y, et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol Int. 2010;60:351–7.
Wang R, Yu Z, Chen F, Xu H, Shen S, Chen W, et al. Mir-300 regulates the epithelial-mesenchymal transition and invasion of hepatocellular carcinoma by targeting the FAK/PI3K/AKT signaling pathway. Biomed Pharmacother. 2018;103:1632.
Acknowledgements
This work was supported by the PhD Start-up Fund of Guangdong Medical University (No.: 2XB17025), Medical Scientific Research Foundation of Guangdong Province of China (No.: B2019024), Project of Traditional Chinese Medicine Bureau of Guangdong Province of China (No.: 20202095) and “Group-type” Special Support Project for Education Talents in Universities (No.: 4SG19045G).
Author information
Authors and Affiliations
Contributions
Guarantor of integrity of the entire study and study concepts: A-HF. Study design: J-LC, D-JL, M-YL, LL. Definition of intellectual content: A-HF, D-JL. Literature research: D-JL, X-YL. Clinical studies: D-JL, M-YL. Experimental studies and data acquisition: J-LC, M-YL. Data analysis and statistical analysis: Y-JP. Manuscript preparation: J-LC, D-JL. Manuscript editing and manuscript review: X-JZ, A-HF.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
The informed consent was obtained from the study participants.
Ethics approval and consent to participate
The protocol has been approved by the Ethics Committee of Guangdong Medical University. All patients were informed of the study and signed the written consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Cheng, JL., Li, DJ., Lv, MY. et al. LncRNA KCNQ1OT1 regulates the invasion and migration of hepatocellular carcinoma by acting on S1PR1 through miR-149. Cancer Gene Ther 28, 850–863 (2021). https://doi.org/10.1038/s41417-020-0203-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41417-020-0203-x
This article is cited by
-
Long non-coding RNA in hepatocellular carcinoma: mechanistic insights and therapeutic perspectives
Cellular Oncology (2025)
-
Sponging of five tumour suppressor miRNAs by lncRNA-KCNQ1OT1 activates BMPR1A/BMPR1B-ACVR2A/ACVR2B signalling and promotes chemoresistance in hepatocellular carcinoma
Cell Death Discovery (2024)
-
An effective N6-methyladenosine-related long non-coding RNA prognostic signature for predicting the prognosis of patients with bladder cancer
BMC Cancer (2021)
-
RETRACTED ARTICLE: H3K27ac-induced lncRNA PAXIP1-AS1 promotes cell proliferation, migration, EMT and apoptosis in ovarian cancer by targeting miR-6744-5p/PCBP2 axis
Journal of Ovarian Research (2021)