Abstract
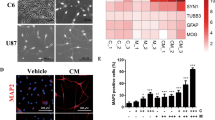
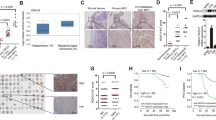
Members of the HDAC family are predictive biomarkers and regulate the tumorigenesis in several cancers. However, the role of these genes in the biology of intracranial ependymomas (EPNs) remains unexplored. Here, an analysis of eighteen HDACs genes in an EPN transcriptomic dataset, revealed significantly higher levels of HDAC4 in supratentorial ZFTA fusion (ST-ZFTA) compared with ST-YAP1 fusion and posterior fossa EPNs, while HDAC7 and SIRT2 were downregulated in ST-ZFTA. HDAC4 was also overexpressed in ST-ZFTA as measured by single-cell RNA-Seq, quantitative real time-polymerase chain reaction, and immunohistochemistry. Survival analyses showed a significantly worse outcome for EPNs with higher HDAC4 and SIRT1 mRNA levels. Ontology enrichment analysis showed an HDAC4-high signature consistent with viral processes while collagen-containing extracellular matrix and cell-cell junction were enriched in those with an HDAC4-low signature. Immune gene analysis demonstrated a correlation between HDAC4 expression and low levels of NK resting cells. Several small molecules compounds targeting HDAC4 and ABCG2, were predicted by in silico analysis to be effective against HDAC4-high ZFTA. Our results provide novel insights into the biology of the HDAC family in intracranial ependymomas and reveal HDAC4 as a prognostic marker and potential therapeutic target in ST-ZFTA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
References
Ostrom QT, Patil N, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro-Oncol. 2020;22:iv1–iv96.
Ellison DW, Aldape KD, Capper D, Fouladi M, Gilbert MR, Gilbertson RJ, et al. cIMPACT‐NOW update 7: advancing the molecular classification of ependymal tumors. Brain Pathol. 2020;30:863–6.
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro-Oncol. 2021;23:1231–51.
Arabzade A, Zhao Y, Varadharajan S, Chen H-C, Jessa S, Rivas B, et al. ZFTA–RELA dictates oncogenic transcriptional programs to drive aggressive supratentorial ependymoma. Cancer Discov. 2021;11:2200–15.
Zheng T, Ghasemi DR, Okonechnikov K, Korshunov A, Sill M, Maass KK, et al. Cross-species genomics reveals oncogenic dependencies in ZFTA/C11orf95 fusion–positive supratentorial ependymomas. Cancer Discov. 2021;11:2230–47.
Andreiuolo F, Varlet P, Tauziède-Espariat A, Jünger ST, Dörner E, Dreschmann V, et al. Childhood supratentorial ependymomas with YAP1-MAMLD1 fusion: an entity with characteristic clinical, radiological, cytogenetic and histopathological features. Brain Pathol. 2019;29:205–16.
Chinnam D, Gupta K, Kiran T, Saraswati A, Salunke P, Madan R, et al. Molecular subgrouping of ependymoma across three anatomic sites and their prognostic implications. Brain Tumor Pathol. 2022;39:151–61.
Kresbach C, Neyazi S, Schüller U. Updates in the classification of ependymal neoplasms: The 2021 WHO Classification and beyond. Brain Pathol. 2022;32:e13068.
Zaytseva M, Papusha L, Novichkova G, Druy A. Molecular stratification of childhood ependymomas as a basis for personalized diagnostics and treatment. Cancers. 2021;13:4954–4954.
Li Y, Seto E. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb Perspect Med. 2016;6:a026831–a026831.
Li G, Tian Y, Zhu W-G. The roles of histone deacetylases and their inhibitors in cancer therapy. Front Cell Dev Biol. 2020;8:576946.
Milde T, Kleber S, Korshunov A, Witt H, Hielscher T, Koch P, et al. A novel human high-risk ependymoma stem cell model reveals the differentiation-inducing potential of the histone deacetylase inhibitor Vorinostat. Acta Neuropathol. 2011;122:637–50.
Antonelli R, Jiménez C, Riley M, Servidei T, Riccardi R, Soriano A, et al. CN133, a novel brain-penetrating histone deacetylase inhibitor, hampers tumor growth in patient-derived pediatric posterior fossa ependymoma models. Cancers. 2020;12:1922–1922.
Pajtler KW, Witt H, Sill M, Jones DTW, Hovestadt V, Kratochwil F, et al. Molecular classification of ependymal tumors across All CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27:728–43.
Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9.
Gillen AE, Riemondy KA, Amani V, Griesinger AM, Gilani A, Venkataraman S, et al. Single-Cell RNA sequencing of childhood ependymoma reveals neoplastic cell subpopulations that impact molecular classification and etiology. Cell Rep. 2020;32:108023–108023.
de Sousa GR, Lira RCP, de Almeida Magalhães T, da Silva KR, Nagano LFP, Saggioro FP, et al. A coordinated approach for the assessment of molecular subgroups in pediatric ependymomas using low-cost methods. J Mol Med. 2021;99:1101–1113.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–8.
Wen Y, Zhang X, Li X, Tian L, Shen S, Ma J, et al. Histone deacetylase (HDAC) 11 inhibits matrix metalloproteinase (MMP) 3 expression to suppress colorectal cancer metastasis. J Cancer. 2022;13:1923–32.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523–1523.
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7.
Merchant TE, Bendel AE, Sabin ND, Burger PC, Shaw DW, Chang E, et al. Conformal radiation therapy for pediatric ependymoma, chemotherapy for incompletely resected ependymoma, and observation for completely resected, supratentorial ependymoma. J Clin Oncol. 2019;37:974–83.
Perla A, Fratini L, Cardoso PS, Nör C, Brunetto AT, Brunetto AL, et al. Histone deacetylase inhibitors in pediatric brain cancers: biological activities and therapeutic potential. Front Cell Dev Biol. 2020;8:546.
Wang Z, Qin G, Zhao TC. HDAC4: mechanism of regulation and biological functions. Epigenomics. 2014;6:139–50.
Mack SC, Pajtler KW, Chavez L, Okonechnikov K, Bertrand KC, Wang XX, et al. Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature. 2018;553:101–5.
Thandapani P. Super-enhancers in cancer. Pharmacol Ther. 2019;199:129–38.
Amodio N, Stamato MA, Gullà AM, Morelli E, Romeo E, Raimondi L, et al. Therapeutic targeting of miR-29b/HDAC4 epigenetic loop in multiple myeloma. Mol Cancer Ther. 2016;15:1364–75.
Cai J-Y, Xu T-T, Wang Y, Chang J-J, Li J, Chen X-Y, et al. Histone deacetylase HDAC4 promotes the proliferation and invasion of glioma cells. Int J Oncol. 2018;53:2758–68.
Zeng L-S, Yang X-Z, Wen Y-F, Mai S-J, Wang M-H, Zhang M-Y, et al. Overexpressed HDAC4 is associated with poor survival and promotes tumor progression in esophageal carcinoma. Aging. 2016;8:1236–48.
Wilson AJ, Byun DS, Nasser S, Murray LB, Ayyanar K, Arango D, et al. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol Biol Cell. 2008;19:4062–75.
Cheng C, Yang J, Li S-W, Huang G, Li C, Min W-P, et al. HDAC4 promotes nasopharyngeal carcinoma progression and serves as a therapeutic target. Cell Death Dis. 2021;12:137–137.
Li J, Yan X, Liang C, Chen H, Liu M, Wu Z, et al. Comprehensive analysis of the differential expression and prognostic value of histone deacetylases in glioma. Front Cell Dev Biol. 2022;10:840759.
Becher OJ. HDAC inhibitors to the rescue in sonic hedgehog medulloblastoma. Neuro Oncol. 2019;21:1091–2.
Lu Y, Stuart JH, Talbot-Cooper C, Agrawal-Singh S, Huntly B, Smid AI, et al. Histone deacetylase 4 promotes type I interferon signaling, restricts DNA viruses, and is degraded via vaccinia virus protein C6. Proc Natl Acad Sci USA. 2019;116:11997–2006.
Yang Q, Tang J, Pei R, Gao X, Guo J, Xu C, et al. Host HDAC4 regulates the antiviral response by inhibiting the phosphorylation of IRF3. J Mol Cell Biol. 2019;11:158–69.
Martini M, Testi MG, Pasetto M, Picchio MC, Innamorati G, Mazzocco M, et al. IFN-gamma-mediated upmodulation of MHC class I expression activates tumor-specific immune response in a mouse model of prostate cancer. Vaccine. 2010;28:3548–57.
Zaidi MR, Merlino G. The two faces of interferon-γ in cancer. Clin Cancer Res. 2011;17:6118–24.
Lee JH, Park SM, Kim OS, Lee CS, Woo JH, Park SJ, et al. Differential SUMOylation of LXRalpha and LXRbeta mediates transrepression of STAT1 inflammatory signaling in IFN-gamma-stimulated brain astrocytes. Mol Cell. 2009;35:806–17.
Zapatka M, Borozan I, Brewer DS, Iskar M, Grundhoff A, Alawi M, et al. The landscape of viral associations in human cancers. Nat Genet. 2020;52:320–30.
Pidugu VK, Pidugu HB, Wu M-M, Liu C-J, Lee T-C. Emerging functions of human IFIT proteins in cancer. Front Mol Biosci. 2019;6:148.
Fensterl V, Sen GC. Interferon-induced ifit proteins: their role in viral pathogenesis. J Virol. 2015;89:2462–8.
Wu S-Y, Fu T, Jiang Y-Z, Shao Z-M. Natural killer cells in cancer biology and therapy. Mol Cancer. 2020;19:120–120.
Mishra R, Welsh R, Szomolanyi-Tsuda E. NK cells and virus-related cancers. Crit Rev Oncog. 2014;19:107–19.
Xu Y-H, Li Z-L, Qiu S-F. IFN-γ induces gastric cancer cell proliferation and metastasis through upregulation of integrin β3-mediated NF-κB signaling. Transl Oncol. 2018;11:182–92.
Ozawa T, Arora S, Szulzewsky F, Juric-Sekhar G, Miyajima Y, Bolouri H, et al. A de novo mouse model of C11orf95-RELA fusion-driven ependymoma identifies driver functions in addition to NF-κB. Cell Rep. 2018;23:3787–97.
Donson AM, Amani V, Warner EA, Griesinger AM, Witt DA, Levy JMM, et al. Identification of FDA-approved oncology drugs with selective potency in high-risk childhood ependymoma. Mol Cancer Ther. 2018;17:1984–94.
Hanson JE, La H, Plise E, Chen Y-H, Ding X, Hanania T, et al. SAHA enhances synaptic function and plasticity in vitro but has limited brain availability in vivo and does not impact cognition. PLoS ONE. 2013;8:e69964–e69964.
Alqudah M, Al‑Samman R, Azaizeh M, Alzoubi K. Amlodipine inhibits proliferation, invasion, and colony formation of breast cancer cells. Biomed Rep. 2022;16:50–50.
Löscher W, Potschka H. Blood-brain barrier active efflux transporters: ATP-binding cassette gene family. NeuroRX. 2005;2:86–98.
Sabnis DH, Storer LCD, Liu J-F, Jackson HK, Kilday JP, Grundy RG, et al. A role for ABCB1 in prognosis, invasion and drug resistance in ependymoma. Sci Rep. 2019;9:10290–10290.
Maklad A, Sharma A, Azimi I. Calcium signaling in brain cancers: roles and therapeutic targeting. Cancers. 2019;11:145–145.
Acknowledgements
We thank Dr. Kristian Pajtler for providing information on the survival clinical data of EPN patients (GSE64415 dataset). We also thank Dr. Maristella B. F. Reis and Dr. Paulo H. S. Klinger for their assistance with the clinical data. Finally, we want to thank and acknowledge patients and families affected by ependymoma for their generous contributions to these studies.
Funding
This study was supported by the São Paulo Research Foundation (FAPESP)- grant no 2014/20341–0, 2018/23372–4/Brazil to G.R.S, 2021/11402–9/Brazil to G.R.S; Brazilian Federal Agency for Support and Evaluation of Graduate Education (CAPES) – Finance Code 001; National Council for Scientific and Technological Development (CNPq), 140140–2019–0. We also had support of FAEPA (Fundação de Amparo ao Ensino, Pesquisa e Assistência do Hospital das Clínicas da Faculdade de Medicina de Ribeirão Preto-USP).
Author information
Authors and Affiliations
Contributions
GRS planned and conducted all experiments, in silico analysis, drafted and wrote, and critically reviewed the manuscript. KBS, PSC, and LCV helped with the bench methodology and investigation and critically read the manuscript. LFPN and KAR performed bioinformatics analysis and interpretation. FPS performed the histopathologic analysis. SKNM, SRB, JAY, IAC, and RGPQ provided reagents, data, consultation, and editing of the manuscript. CAS, AMD, NKF, ETV, and LGT provided the consultation review and editing of the manuscript. All authors critically wrote, edited, and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This research was submitted to and approved by the HC/FMRP-USP Research Ethics Committee (CAAE no 40768820.4.0000.5440).
Informed consent
All samples were obtained after receiving informed consent from all participant´s legal guardians included in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
de Sousa, G.R., Salomão, K.B., Nagano, L.F.P. et al. Identification of HDAC4 as a potential therapeutic target and prognostic biomarker for ZFTA-fused ependymomas. Cancer Gene Ther 30, 1105–1113 (2023). https://doi.org/10.1038/s41417-023-00616-z
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41417-023-00616-z