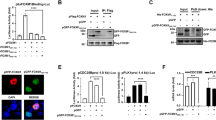
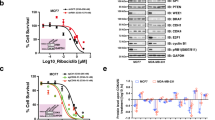
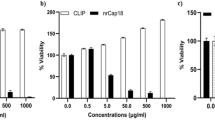
Abstract
CDK1 is an oncogenic serine/threonine kinase known to play an important role in the regulation of the cell cycle. FOXM1, as one of the CDK1 substrates, requires binding of CDK1/CCNB1 complex for phosphorylation-dependent recruitment of p300/CBP coactivators to mediate transcriptional activity. Previous studies from our laboratory found that a novel peptide (M1-20) derived from the C-terminus of FOXM1 exhibited potent inhibitory effects for cancer cells. Based on these proofs and to explore the inhibitory mechanism of M1-20, we designed experiments and found that CDK1 served as an important target of M1-20. M1-20 enhanced the ubiquitination and degradation of CDK1 by CUL4-DDB1-DCAF1 complexes through the proteasome pathway. M1-20 could also affect the formation of CDK1/CCNB1 complexes. In addition, compared to RO3306, a CDK1 inhibitor, M1-20 exhibited excellent inhibitory effects in FVB/N MMTV-PyVT murine model of spontaneous breast cancer. These results suggested that M1-20 was a potential CDK1 inhibitor for the treatment of cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this article and its Supplementary Information files. Additional data are available from the corresponding author on reasonable request.
References
Banyai G, Baidi F, Coudreuse D, Szilagyi Z. Cdk1 activity acts as a quantitative platform for coordinating cell cycle progression with periodic transcription. Nat Commun. 2016;7:11161.
Campsteijn C, Ovrebo JI, Karlsen BO, Thompson EM. Expansion of cyclin D and CDK1 paralogs in Oikopleura dioica, a chordate employing diverse cell cycle variants. Mol Biol Evol. 2012;29:487–502.
Allan LA, Clarke PR. Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol Cell. 2007;26:301–10.
Neganova I, Tilgner K, Buskin A, Paraskevopoulou I, Atkinson SP, Peberdy D, et al. CDK1 plays an important role in the maintenance of pluripotency and genomic stability in human pluripotent stem cells. Cell Death Dis. 2014;5:e1508.
Michowski W, Chick JM, Chu C, Kolodziejczyk A, Wang Y, Suski JM, et al. Cdk1 controls global epigenetic landscape in embryonic stem cells. Mol Cell. 2020;78:459–76.e13.
Izadi S, Nikkhoo A, Hojjat-Farsangi M, Namdar A, Azizi G, Mohammadi H, et al. CDK1 in breast cancer: implications for theranostic potential. Anticancer Agents Med Chem. 2020;20:758–67.
Xi Q, Huang M, Wang Y, Zhong J, Liu R, Xu G, et al. The expression of CDK1 is associated with proliferation and can be a prognostic factor in epithelial ovarian cancer. Tumour Biol. 2015;36:4939–48.
Huang J, Chen P, Liu K, Liu J, Zhou B, Wu R, et al. CDK1/2/5 inhibition overcomes IFNG-mediated adaptive immune resistance in pancreatic cancer. Gut. 2021;70:890–9.
Chen X, Zhang FH, Chen QE, Wang YY, Wang YL, He JC, et al. The clinical significance of CDK1 expression in oral squamous cell carcinoma. Med Oral Patol Oral Cir Bucal. 2015;20:e7–12.
Huang Z, Shen G, Gao J. CDK1 promotes the stemness of lung cancer cells through interacting with Sox2. Clin Transl Oncol. 2021;23:1743–51.
Ravindran Menon D, Luo Y, Arcaroli JJ, Liu S, KrishnanKutty LN, Osborne DG, et al. CDK1 interacts with Sox2 and promotes tumor initiation in human melanoma. Cancer Res. 2018;78:6561–74.
Nagy A, Munkacsy G, Gyorffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021;11:6047.
Kim SJ, Nakayama S, Miyoshi Y, Taguchi T, Tamaki Y, Matsushima T, et al. Determination of the specific activity of CDK1 and CDK2 as a novel prognostic indicator for early breast cancer. Ann Oncol. 2008;19:68–72.
Xia Q, Cai Y, Peng R, Wu G, Shi Y, Jiang W. The CDK1 inhibitor RO3306 improves the response of BRCA-pro fi cient breast cancer cells to PARP inhibition. Int J Oncol. 2014;44:735–44.
Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA. 2006;103:10660–5.
Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115.
Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–5.
Aleem E, Kiyokawa H, Kaldis P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat Cell Biol. 2005;7:831–6.
Welch PJ, Wang JY. Coordinated synthesis and degradation of cdc2 in the mammalian cell cycle. Proc Natl Acad Sci USA. 1992;89:3093–7.
Yoon CH, Miah MA, Kim KP, Bae YS. New Cdc2 Tyr 4 phosphorylation by dsRNA-activated protein kinase triggers Cdc2 polyubiquitination and G2 arrest under genotoxic stresses. EMBO Rep. 2010;11:393–9.
Herrero-Ruiz J, Mora-Santos M, Giraldez S, Saez C, Japon MA, Tortolero M, et al. betaTrCP controls the lysosome-mediated degradation of CDK1, whose accumulation correlates with tumor malignancy. Oncotarget. 2014;5:7563–74.
Bu H, Lan X, Cheng H, Pei C, Ouyang M, Chen Y, et al. Development of an interfering peptide M1-20 with potent anti-cancer effects by targeting FOXM1. Cell Death Dis. 2023;14:533.
Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–61.
Raveh B, London N, Schueler-Furman O. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins-Struct Funct Bioinforma. 2010;78:2029–40.
Barlow KA, Ó Conchúir S, Thompson S, Suresh P, Lucas JE, Heinonen M, et al. Flex ddG: Rosetta ensemble-based estimation of changes in protein-protein binding affinity upon mutation. J Phys Chem B. 2018;122:5389–99.
Zhang Z, Bu H, Yu J, Chen Y, Pei C, Yu L, et al. The cell-penetrating FOXM1 N-terminus (M1-138) demonstrates potent inhibitory effects on cancer cells by targeting FOXM1 and FOXM1-interacting factor SMAD3. Theranostics. 2019;9:2882–96.
Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–61.
Stranges PB, Kuhlman B. A comparison of successful and failed protein interface designs highlights the challenges of designing buried hydrogen bonds. Protein Sci. 2013;22:74–82.
Shi Z, Tian L, Qiang T, Li J, Xing Y, Ren X, et al. From structure modification to drug launch: a systematic review of the ongoing development of cyclin-dependent kinase inhibitors for multiple cancer therapy. J Med Chem. 2022;65:6390–418.
Schneider-Poetsch T, Ju JH, Eyler DE, Dang YJ, Bhat S, Merrick WC, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–17.
Said A, Bock S, Lajqi T, Müller G, Weindl G. Chloroquine promotes IL-17 production by CD4 T cells via p38-Dependent IL-23 release by monocyte-derived langerhans-like cells. J Immunol. 2014;193:6135–43.
MacLaren AP, Chapman RS, Wyllie AH, Watson CJ. p53-dependent apoptosis induced by proteasome inhibition in mammary epithelial cells. Cell Death Differ. 2001;8:210–8.
Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220.
Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–U67.
Harper JW, Schulman BA. Cullin-RING ubiquitin ligase regulatory circuits: a quarter century beyond the F-Box hypothesis. Annu Rev Biochem. 2021;90:403–29.
Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–70.
Lee J, Zhou PB. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell. 2007;26:775–80.
Wang X, Arceci A, Bird K, Mills CA, Choudhury R, Kernan JL, et al. VprBP/DCAF1 regulates the degradation and nonproteolytic activation of the cell cycle transcription factor FoxM1. Mol Cell Biol. 2017;37:e00609–16.
Fourest-Lieuvin A, Peris L, Gache V, Garcia-Saez I, Juillan-Binard C, Lantez V, et al. Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol Biol Cell. 2006;17:1041–50.
Serpico AF, Febbraro F, Pisauro C, Grieco D. Compartmentalized control of Cdk1 drives mitotic spindle assembly. Cell Rep. 2022;38:110305.
Attalla S, Taifour T, Bui T, Muller W. Insights from transgenic mouse models of PyMT-induced breast cancer: recapitulating human breast cancer progression in vivo. Oncogene. 2021;40:475–91.
Arslan C, Altundag K, Dizdar O. Emerging drugs in metastatic breast cancer: an update. Expert Opin Emerg Dr. 2011;16:647–67.
Wang Q, Bode AM, Zhang T. Targeting CDK1 in cancer: mechanisms and implications. NPJ Precis Oncol. 2023;7:58.
Massacci G, Perfetto L, Sacco F. The Cyclin-dependent kinase 1: more than a cell cycle regulator. Br J Cancer. 2023;129:1707–16.
Garcia-Sampedro A, Gaggia G, Ney A, Mahamed I, Acedo P. The state-of-the-art of Phase II/III clinical trials for targeted pancreatic cancer therapies. J Clin Med. 2021;10:566.
Wu CX, Wang XQ, Chok SH, Man K, Tsang SHY, Chan ACY, et al. Blocking CDK1/PDK1/beta-Catenin signaling by CDK1 inhibitor RO3306 increased the efficacy of sorafenib treatment by targeting cancer stem cells in a preclinical model of hepatocellular carcinoma. Theranostics. 2018;8:3737–50.
Han Z, Jia Q, Zhang J, Chen M, Wang L, Tong K, et al. Deubiquitylase YOD1 regulates CDK1 stability and drives triple-negative breast cancer tumorigenesis. J Exp Clin Cancer Res. 2023;42:228.
Ci MX, Zhao GC, Li CY, Liu RC, Hu XS, Pan J, et al. OTUD4 promotes the progression of glioblastoma by deubiquitinating CDK1 and activating MAPK signaling pathway. Cell Death Dis. 2024;15:179.
Suski JM, Braun M, Strmiska V, Sicinski P. Targeting cell-cycle machinery in cancer. Cancer Cell. 2021;39:759–78.
Békés M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 2022;21:181–200.
Attwood MM, Fabbro D, Sokolov AV, Knapp S, Schiöth HB. Trends in kinase drug discovery: targets, indications and inhibitor design. Nat Rev Drug Discov. 2021;20:839–61.
Wong RL, Choi MY, Wang HY, Kipps TJ. Mutation in Bruton Tyrosine Kinase (BTK) A428D confers resistance To BTK-degrader therapy in chronic lymphocytic leukemia. Leukemia. 2024;38:1818–21.
Domostegui A, Nieto-Barrado L, Perez-Lopez C, Mayor-Ruiz C. Chasing molecular glue degraders: screening approaches. Chem Soc Rev. 2022;51:5498–517.
Alvarez-Fernandez M, Malumbres M. Mechanisms of sensitivity and resistance to CDK4/6 Inhibition. Cancer Cell. 2020;37:514–29.
Olson CM, Liang Y, Leggett A, Park WD, Li L, Mills CE, et al. Development of a selective CDK7 covalent inhibitor reveals predominant cell-cycle phenotype. Cell Chem Biol. 2019;26:792–803.e10.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 81773169, No. 81472718), Hunan Key R&D Project (2023SK2040), China Changsha Development and Reform Commission “Mass entrepreneurship and innovation program” (2018-68) and “Innovation platform construction program” (2018-216), Hunan Natural Science Foundation (812202201381).
Author information
Authors and Affiliations
Contributions
YT designed the study. HB performed the most experiments and analyzed the data. CP and MO contributed to data acquisition or analysis. YT, LY, and XH provided support with experiments. YT and HB wrote and edited the manuscript. YT, LY, XH, and YC supervised the project, and YT performed project administration and funding acquisition.
Corresponding author
Ethics declarations
Competing interests
A patent on M1-20 and its derivatives has been granted in China (ZL202011200439.0). Y. T. and H. B. are co-inventors of this patent. This does not alter the authors′ adherence to the policies on sharing data and materials.
Ethics approval
All animal care and experiments were performed in accordance with guidelines, approved by the Laboratory Animal Center of Hunan, China (Protocol No. SYXK [Xiang] 2018-0006).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bu, H., Pei, C., Ouyang, M. et al. The antitumor peptide M1-20 induced the degradation of CDK1 through CUL4-DDB1-DCAF1-involved ubiquitination. Cancer Gene Ther 32, 61–70 (2025). https://doi.org/10.1038/s41417-024-00855-8
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41417-024-00855-8
This article is cited by
-
Kaempferol triggers cellular senescence via CDK1 ubiquitination in HCC cells
Cancer Cell International (2025)