Abstract
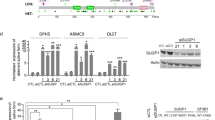
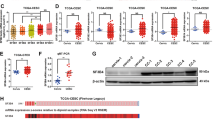
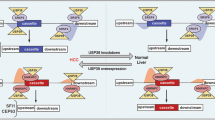
Splicing factor 3B (SF3B) subunit 4 (SF3B4), an SF3B complex component essential for spliceosome assembly and accurate splicing, plays a major role in cancer development. However, the precise mechanism through which SF3B4 contributes to tumor growth remains unclear. Here, we demonstrate that SF3B4 is strongly expressed in patients with various cancer types and correlated with their survival. By using hepatocellular carcinoma (HCC) as a model, we reveal that SF3B4’s interactions with and regulatory influence on the checkpoint protein BUB1 are essential for appropriate cancer cell mitosis and proliferation. Our results thus demonstrate the roles of SF3B4 as both a cell-cycle regulator and an oncogenic factor in HCC, highlighting its potential as a pan-cancer therapeutic target and diagnostic biomarker.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The datasets used in this study are available from the corresponding author upon reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD052853.
References
The global challenge of cancer. Nat Cancer. 2020;1:1–2.
Lancet Public The. H. Inequalities in cancer: a major public health concern. Lancet Public Health. 2024;9:e147.
He S, Xia C, Li H, Cao M, Yang F, Yan X, et al. Cancer profiles in China and comparisons with the USA:a comprehensive analysis in the incidence,mortality,survival,staging,and attribution to risk factors. Science China. 2024;67:122–31.
Ding R, Yu X, Hu Z, Dong Y, Huang H, Zhang Y, et al. Lactate modulates RNA splicing to promote CTLA-4 expression in tumor-infiltrating regulatory T cells. Immunity. 2024;57:528–40.e6.
Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18.
Mm G, Sander B, Cl W, Luhrmann R, Stark H. Molecular Architecture of the Multiprotein Splicing Factor SF3b. Science. 2003;300:980–84.
Bland P, Saville H, Wai PT, Curnow L, Muirhead G, Nieminuszczy J, et al. SF3B1 hotspot mutations confer sensitivity to PARP inhibition by eliciting a defective replication stress response. Nat Genet. 2023;55:1311–23.
Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nature Rev Cancer. 2016;16:413–30.
O’Connor CM, Narla G. Splice of Life for Cancer: Missplicing of PPP2R5A by Mutant SF3B1 Leads to MYC Stabilization and Tumorigenesis. Cancer Discov. 2020;10:765–67.
Patnaik MM. How I diagnose and treat chronic myelomonocytic leukemia. Haematologica. 2022;107:1503–17.
Wang S, Liu Y, Xiao H, Chen Z, Yang X, Yin J, et al. Inhibition of SF3B1 improves the immune microenvironment through pyroptosis and synergizes with alphaPDL1 in ovarian cancer. Cell Death Dis. 2023;14:775.
Gangat N, Patnaik MM, Tefferi A. Myelodysplastic syndromes: Contemporary review and how we treat. Am J Hematol. 2016;91:76–89.
Steensma DP, Wermke M, Klimek VM, Greenberg PL, Platzbecker U. Phase I First-in-Human Dose Escalation Study of the oral SF3B1 modulator H3B-8800 in myeloid neoplasms. Leukemia. 2021;35:3542–50.
Sciarrillo R, Wojtuszkiewicz A, Assaraf YG, Jansen G, Kaspers GJL, Giovannetti E, et al. The role of alternative splicing in cancer: From oncogenesis to drug resistance. Drug Resist Updat. 2020;53:100728.
Wang SJ, Dougan SK, Dougan M. Immune mechanisms of toxicity from checkpoint inhibitors. Trends cancer. 2023;9:543–53.
Larsen NA. The SF3b Complex is an Integral Component of the Spliceosome and Targeted by Natural Product-Based Inhibitors. Subcell Biochem. 2021;96:409–32.
Champion-Arnaud P, Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 1994;8:1974–83.
Bernier FP, Caluseriu O, Ng S, Schwartzentruber J, Buckingham KJ, Innes AM, et al. Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet. 2012;90:925–33.
Diao Y, Li Y, Wang Z, Wang S, Li P, Kong B. SF3B4 promotes ovarian cancer progression by regulating alternative splicing of RAD52. Cell Death Dis. 2022;13:179.
Li Y, Diao Y, Wang Z, Wang S, Peng J, Kong B. The splicing factor SF3B4 drives proliferation and invasion in cervical cancer by regulating SPAG5. Cell Death Discov. 2022;8:326.
Liu Z, Li W, Pang Y, Zhou Z, Liu S, Cheng K, et al. SF3B4 is regulated by microRNA-133b and promotes cell proliferation and metastasis in hepatocellular carcinoma. EBioMedicine. 2018;38:57–68.
Han C, Chen J, Huang J, Zhu R, Zeng J, Yu H, et al. Single-cell transcriptome analysis reveals the metabolic changes and the prognostic value of malignant hepatocyte subpopulations and predict new therapeutic agents for hepatocellular carcinoma. Front Oncol. 2023;13:1104262.
Shen Q, Nam SW. SF3B4 as an early-stage diagnostic marker and driver of hepatocellular carcinoma. BMB Rep. 2018;51:57–8.
Caicedo HH, Hashimoto DA, Caicedo JC, Pentland A, Pisano GP. Visualizing and interpreting cancer genomics data via the Xena platform. Nature Biotechnol. 2020;38:669–73.
Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D63.
Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, et al. A subcellular map of the human proteome. Science. 2017; 356:eaal3321.
Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900–D08.
Shen W, Song Z, Zhong X, Huang M, Shen D, Gao P, et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. Imeta. 2022;1:e36.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7.
Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
Pan Q, Luo P, Shi C. PC4-mediated Ku complex PARylation facilitates NHEJ-dependent DNA damage repair. J Biol Chem. 2023;299:105032.
Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303–22.
Li X, Ma S, Deng Y, Yi P, Yu J. Targeting the RNA m(6)A modification for cancer immunotherapy. Mol Cancer. 2022;21:76.
Liu X, Du Y, Huang Z, Qin H, Chen J, Zhao Y. Insights into roles of METTL14 in tumors. Cell Prolif. 2022;55:e13168.
Global cancer burden growing, amidst mounting need for services. Saudi Medical Journal. 2024; 45:326–7.
Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res. 2021;149:1–61.
Singal AG, Kanwal F, Llovet JM. Global trends in hepatocellular carcinoma epidemiology: implications for screening, prevention and therapy. Nat Rev Clin Oncol. 2023;20:864–84.
Llovet JM, Pinyol R, Kelley RK, El-Khoueiry A, Reeves HL, Wang XW, et al. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat Cancer. 2022;3:386–401.
Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells?. Cell Stem Cell. 2015;16:225–38.
Kreso A, O’Brien CA, van Galen P, Gan OI, Notta F, Brown AM, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–8.
Fischer M, Schade AE, Branigan TB, Müller GA, DeCaprio JA. Coordinating gene expression during the cell cycle. Trends Biochemical Sci. 2022;47:1009–22.
Gallo D, Young JTF, Fourtounis J, Martino G, Álvarez-Quilón A, Bernier C, et al. CCNE1 amplification is synthetic lethal with PKMYT1 kinase inhibition. Nature. 2022;604:749–56.
Sher G, Masoodi T, Patil K, Akhtar S, Kuttikrishnan S, Ahmad A, et al. Dysregulated FOXM1 signaling in the regulation of cancer stem cells. Semin Cancer Biol. 2022;86:107–21.
Elowe S, Bolanos-Garcia VM. The spindle checkpoint proteins BUB1 and BUBR1: (SLiM)ming down to the basics. Trends Biochemical Sci. 2022;47:352–66.
Singh P, Pesenti ME, Maffini S, Carmignani S, Hedtfeld M, Petrovic A, et al. BUB1 and CENP-U, Primed by CDK1, Are the Main PLK1 Kinetochore Receptors in Mitosis. Mol Cell. 2021;81:67–87.e9.
Wang X, Babu JR, Harden JM, Jablonski SA, Gazi MH, Lingle WL, et al. The Mitotic Checkpoint Protein hBUB3 and the mRNA Export Factor hRAE1 Interact with GLE2p-binding Sequence (GLEBS)-containing Proteins*. J Biol Chem. 2001;276:26559–67.
Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11.
Ding J, Li C, Cheng Y, Du Z, Wang Q, Tang Z, et al. Alterations of RNA splicing patterns in esophagus squamous cell carcinoma. Cell Biosci. 2021;11:36.
He G, Gu K, Wei J, Zhang J. METTL3-mediated the m6A modification of SF3B4 facilitates the development of non-small cell lung cancer by enhancing LSM4 expression. Thorac Cancer. 2024;15:919–28.
Iguchi T, Masuda T, Nambara S, Kidogami S, Ogawa Y, Hu Q, et al. Increased Copy Number of the Gene Encoding SF3B4 Indicates Poor Prognosis in Hepatocellular Carcinoma. Anticancer Res. 2016;36:2139–44.
Lu SX, De Neef E, Thomas JD, Sabio E, Rousseau B, Gigoux M, et al. Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell. 2021;184:4032–47.e31.
Venkataramany AS, Schieffer KM, Lee K, Cottrell CE, Wang PY, Mardis ER, et al. Alternative RNA splicing defects in pediatric cancers: new insights in tumorigenesis and potential therapeutic vulnerabilities. Ann Oncol. 2022;33:578–92.
Acknowledgements
We thank Professor Shi Chunmeng from the Affiliated Hospital of the Third Military Medical University for his discussion regarding the manuscript. Mass spectrometry analysis was performed by the Bioinformatics and Omics Center, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University.
Funding
This work was supported by grants from the National Natural Science Foundation of China [grant numbers 82373023, 82273033, 82072924]; the Guangdong Science and Technology Department [grant number 2022B1515020100]; and the Guangzhou Bureau of Science and Technology [grant number 202201020575, 2024A04J4691].
Author information
Authors and Affiliations
Contributions
The conceptualization and writing of this manuscript were performed by YS and QP. The methodology was designed and implemented by YS, QP, and WC. Data validation and quality control were conducted by LX. Specimen collection and processing were carried out by ST. Literature review and comprehensive analysis were completed by ZY and MZ. Manuscript revision and supervised were performed by DY, LL and JL.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval and consent to participate
All methods in this study were performed in accordance with the relevant guidelines and regulations. Ethical approval for this study was obtained from the Sun Yat-sen Memorial Hospital, Sun Yat-Sen University and adhered to the principles of the Declaration of Helsink. Approval No. SYSKY-2023-010-01. Informed consent was obtained from all human participants prior to their inclusion in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, Y., Pan, Q., Chen, W. et al. Pan-cancer oncogenic properties and therapeutic potential of SF3B4. Cancer Gene Ther 32, 706–720 (2025). https://doi.org/10.1038/s41417-025-00910-y
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41417-025-00910-y